CRISPR is a way off being using in human treatment – but a new discovery could unlock its potential. Here’s what’s new.



This level of precision could be a game-changer for therapies that require gene expression in one specific tissue, without impacting others.
By providing more control over where and when genes are activated, these AI-designed CREs could potentially be used in a variety of therapeutic applications, from treating genetic diseases to optimizing tissue regeneration.
As this AI-powered approach to designing CREs matures, the possibilities are vast. Beyond basic research, these synthetic DNA switches could be employed in biomanufacturing or to develop advanced treatments for a range of conditions, offering more effective ways to manipulate genes with unprecedented precision.

The acetate would then be used to feed plants that are grown hydroponically. The method could also be used to grow other food-producing organisms, since acetate is naturally used by mushrooms, yeast, and algae.
“The whole point of this new process to try to boost the efficiency of photosynthesis,” says senior author Feng Jiao, an electrochemist at Washington University in St. Louis. “Right now, we are at about 4% efficiency, which is already four times higher than for photosynthesis, and because everything is more efficient with this method, the CO2 footprint associated with the production of food becomes much smaller.”
To genetically engineer acetate-eating plants, the researchers are taking advantage of a metabolic pathway that germinating plants use to break down food stored in their seeds. This pathway is switched off once plants become capable of photosynthesis, but switching it back on would enable them to use acetate as a source of energy and carbon.
Donate to Closer To Truth and help us keep our content free and without paywalls: https://shorturl.at/OnyRq.
What is information in biology? information is essential for analyzing data and testing hypotheses. But what is information in evolution, population genetics, levels of selection, and molecular genetics? Is computational biology transformational?
Follow Closer To Truth on Instagram for news, announcements, and exciting updates: https://shorturl.at/p2IhM
Terrence William Deacon is an American neuroanthropologist. He taught at Harvard for eight years, relocated to Boston University in 1992, and is currently Professor of Anthropology and member of the Cognitive Science Faculty at the University of California, Berkeley.
Get member exclusives like early access to new content with a free Closer To Truth account: https://closertotruth.com/
Closer To Truth, hosted by Robert Lawrence Kuhn and directed by Peter Getzels, presents the world’s greatest thinkers exploring humanity’s deepest questions. Discover fundamental issues of existence. Engage new and diverse ways of thinking. Appreciate intense debates. Share your own opinions. Seek your own answers.
Michael Levin is a Distinguished Professor in the Biology department at Tufts University and associate faculty at the Wyss Institute for Bioinspired Engineering at Harvard University. @drmichaellevin holds the Vannevar Bush endowed Chair and serves as director of the Allen Discovery Center at Tufts and the Tufts Center for Regenerative and Developmental Biology. Prior to college, Michael Levin worked as a software engineer and independent contractor in the field of scientific computing. He attended Tufts University, interested in artificial intelligence and unconventional computation. To explore the algorithms by which the biological world implemented complex adaptive behavior, he got dual B.S. degrees, in CS and in Biology and then received a PhD from Harvard University. He did post-doctoral training at Harvard Medical School, where he began to uncover a new bioelectric language by which cells coordinate their activity during embryogenesis. His independent laboratory develops new molecular-genetic and conceptual tools to probe large-scale information processing in regeneration, embryogenesis, and cancer suppression.
TIMESTAMPS:
0:00 — Introduction.
1:41 — Creating High-level General Intelligences.
7:00 — Ethical implications of Diverse Intelligence beyond AI & LLMs.
10:30 — Solving the Fundamental Paradox that faces all Species.
15:00 — Evolution creates Problem Solving Agents & the Self is a Dynamical Construct.
23:00 — Mike on Stephen Grossberg.
26:20 — A Formal Definition of Diverse Intelligence (DI)
30:50 — Intimate relationships with AI? Importance of Cognitive Light Cones.
38:00 — Cyborgs, hybrids, chimeras, & a new concept called “Synthbiosis“
45:51 — Importance of the symbiotic relationship between Science & Philosophy.
53:00 — The Space of Possible Minds.
58:30 — Is Mike Playing God?
1:02:45 — A path forward: through the ethics filter for civilization.
1:09:00 — Mike on Daniel Dennett (RIP)
1:14:02 — An Ethical Synthbiosis that goes beyond “are you real or faking it“
1:25:47 — Conclusion.
EPISODE LINKS:
- Mike’s Round 1: https://youtu.be/v6gp-ORTBlU
- Mike’s Round 2: https://youtu.be/kMxTS7eKkNM
- Mike’s Channel: https://www.youtube.com/@drmichaellevin.
- Mike’s Website: https://drmichaellevin.org/
- Blog Website: https://thoughtforms.life.
- Mike’s Twitter: https://twitter.com/drmichaellevin.
- Mike’s Publications: https://scholar.google.com/citations?user=luouyakAAAAJ&hl=en.
- Mike’s NOEMA piece: https://www.noemamag.com/ai-could-be-a-bridge-toward-diverse-intelligence/
- Stephen Grossberg: https://youtu.be/bcV1eSgByzg.
- Mark Solms: https://youtu.be/rkbeaxjAZm4
- VPRO Roundtable: https://youtu.be/RVrnn7QW6Jg?feature=shared.
CONNECT:
- Website: https://tevinnaidu.com.
- Podcast: https://podcasters.spotify.com/pod/show/drtevinnaidu.
- Twitter: https://twitter.com/drtevinnaidu.
- Facebook: https://www.facebook.com/drtevinnaidu.
- Instagram: https://www.instagram.com/drtevinnaidu.
- LinkedIn: https://www.linkedin.com/in/drtevinnaidu.
Disclaimer: The information provided on this channel is for educational purposes only. The content is shared in the spirit of open discourse and does not constitute, nor does it substitute, professional or medical advice. We do not accept any liability for any loss or damage incurred from you acting or not acting as a result of listening/watching any of our contents. You acknowledge that you use the information provided at your own risk. Listeners/viewers are advised to conduct their own research and consult with their own experts in the respective fields.
#MichaelLevin #DiverseIntelligence #AI #Mind
Researchers at The Jackson Laboratory (JAX), the Broad Institute of MIT and Harvard, and Yale University, have used artificial intelligence to design thousands of new DNA switches that can precisely control the expression of a gene in different cell types. Their new approach opens the possibility of controlling when and where genes are expressed in the body, for the benefit of human health and medical research, in ways never before possible.
“What is special about these synthetically designed elements is that they show remarkable specificity to the target cell type they were designed for,” said Ryan Tewhey, PhD, an associate professor at The Jackson Laboratory and co-senior author of the work. “This creates the opportunity for us to turn the expression of a gene up or down in just one tissue without affecting the rest of the body.”
In recent years, genetic editing technologies and other gene therapy approaches have given scientists the ability to alter the genes inside living cells. However, affecting genes only in selected cell types or tissues, rather than across an entire organism, has been difficult. That is in part because of the ongoing challenge of understanding the DNA switches, called cis-regulatory elements (CREs), that control the expression and repression of genes.
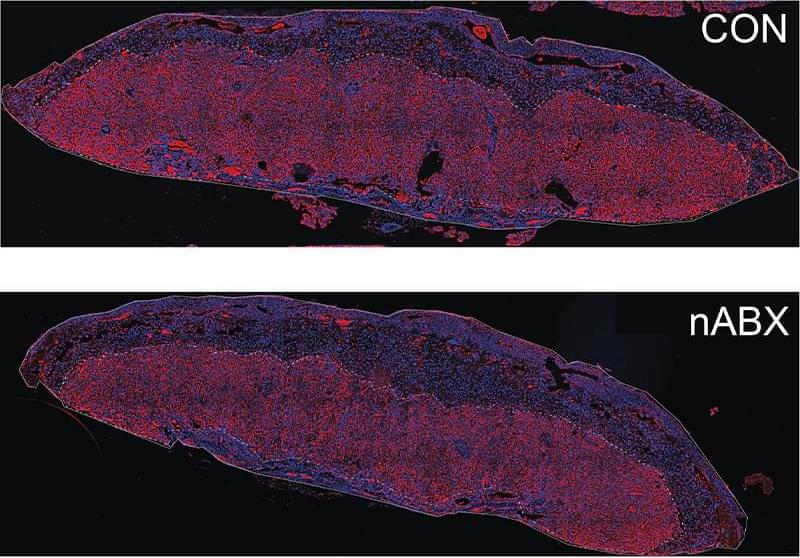
ABOVE: The placenta’s labyrinth zone (red), responsible for nutrient exchange between mother and fetus, is reduced in fetuses with dysbiotic fathers (lower panel) compared to healthy fathers (upper panel). Ayele Argaw-Denboba.
The microbiome has a profound impact on host health that extends to the host’s young ones. Studies in mice have shown that maternal gut bacteria play a role in offspring behavior and placental growth during pregnancy.1,2 Yet, the effects of the paternal microbiome on the health of their progeny remained relatively unexplored.
In a new study, scientists found that altering the gut microbiome of male mice negatively affected the health and lifespan of their offspring through epigenetic changes in the sperm.3 The results, published in Nature, offer insights into a gut-germline axis that mediates the effects of the microbiome on health and disease across generations.

In the consequent tweets, the biohacker attributed his successful hair regeneration to a multi-faceted approach. The key to his transformation has been the strategic use of vitamins and nutrients, particularly protein and Omega-3 fatty acids, which have played a crucial role in restoring his hair.
In addition to nutrition, he has developed a personalised topical formula tailored to his genetics, that includes melatonin, caffeine, and Vitamin D3. He has also incorporated red light therapy into his daily routine, even wearing a specialised hat to administer this treatment throughout the day.
Another critical component of Johnson’s regimen is oral minoxidil, a topical hair-loss drug. However, he stressed that it is only considered safe at low doses as it can lead to unpleasant side effects, including excessive hair growth and headaches.
BIOHACKER Bryan Johnson who’s shelling out millions in his quest for immortality has revealed the exact steps he follows to reverse his hair loss and get rid of greys.
The tech-tycoon, 47, claimed he’s been able to grow a full head of hair despite being “genetically bald” through a mix of supplements, red light therapy and customised hair oil.
At one time, Bryan was best known for founding the payments company Braintree — but nowadays he’s making headlines for his very expensive quest to turn back the clock and become 18 again.

The reason? While sunny regions naturally provide enough light to grow crops, areas with colder winters often need grow lights and greenhouses part of the year. This increases energy consumption, logistical headaches, and ultimately, food costs.
In their paper, Jiao and colleagues argue for a new method that could dramatically revamp farming practices to reduce land use and greenhouse gas emissions.
Dubbed “electro-agriculture,” the approach uses solar panels to trigger a chemical reaction that turns ambient CO2 into an energy source called acetate. Certain mushrooms, yeast, and algae already consume acetate as food. With a slight genetic tweak, we could also engineer other common foods such as grains, tomatoes, or lettuce to consume acetate.