Learn more about the cutting-edge technology that enables 3D printing inside living cells and what that means for the bioengineering of tomorrow.
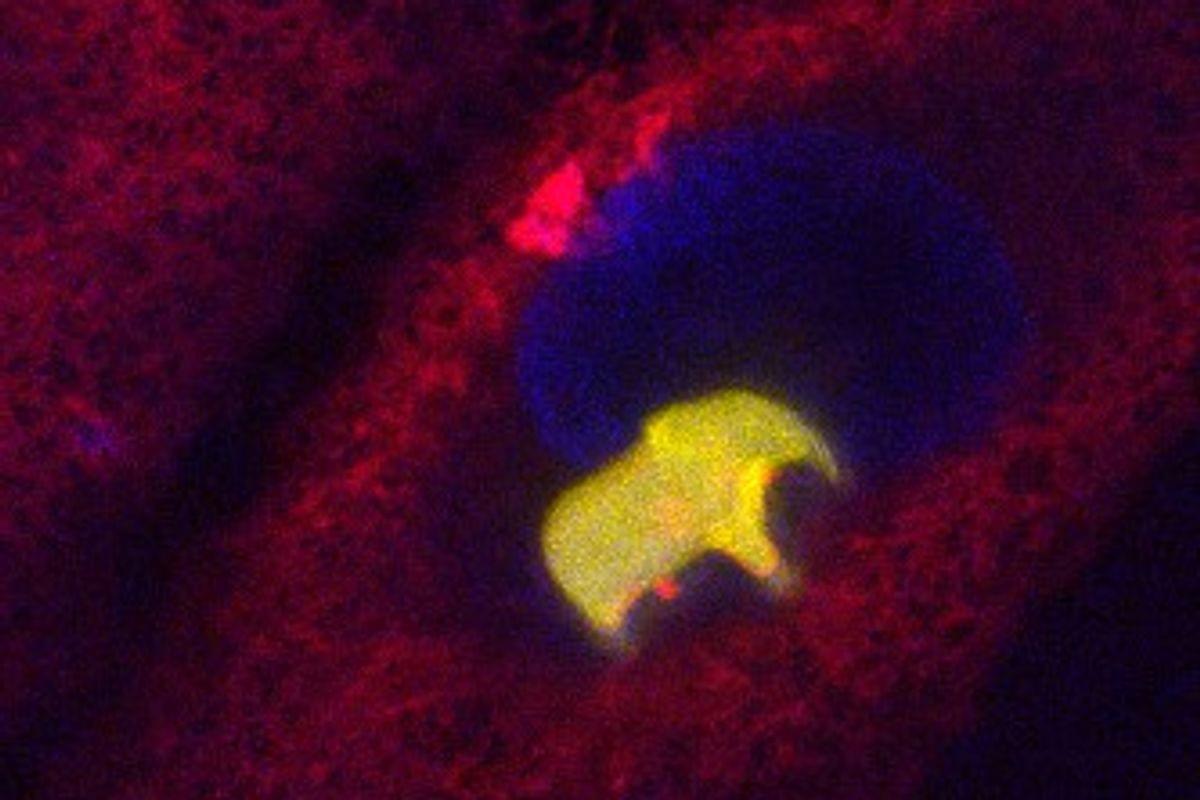

Depending on others for something you need may feel like a risky proposition—and perhaps a human one. It is actually a survival strategy found in the microbial world, and far more frequently than one might expect. Discovering why is key to understanding how microbes form stable communities across medical, industrial, and ecological settings.
A new study by bioengineering professor Sergei Maslov (CAIM co-leader), computational scientist Ashish George, and biology professor Tong Wang explores why interdependence can be such a winning move for microbial communities. Their work, published in Cell Systems, demonstrated that a mathematical model of how bacteria produce and share resources accurately predicted the outcome of experiments with living E. coli strains.
The researchers’ collaboration began during their time as colleagues at the Carl R. Woese Institute for Genomic Biology at the University of Illinois Urbana-Champaign. George continued the collaboration in his position at the Broad Institute; Wang, in his appointment at Purdue University. Maslov, who led the study, remains at Illinois and is an affiliate member of the National Institute for Theory and Mathematics in Biology.
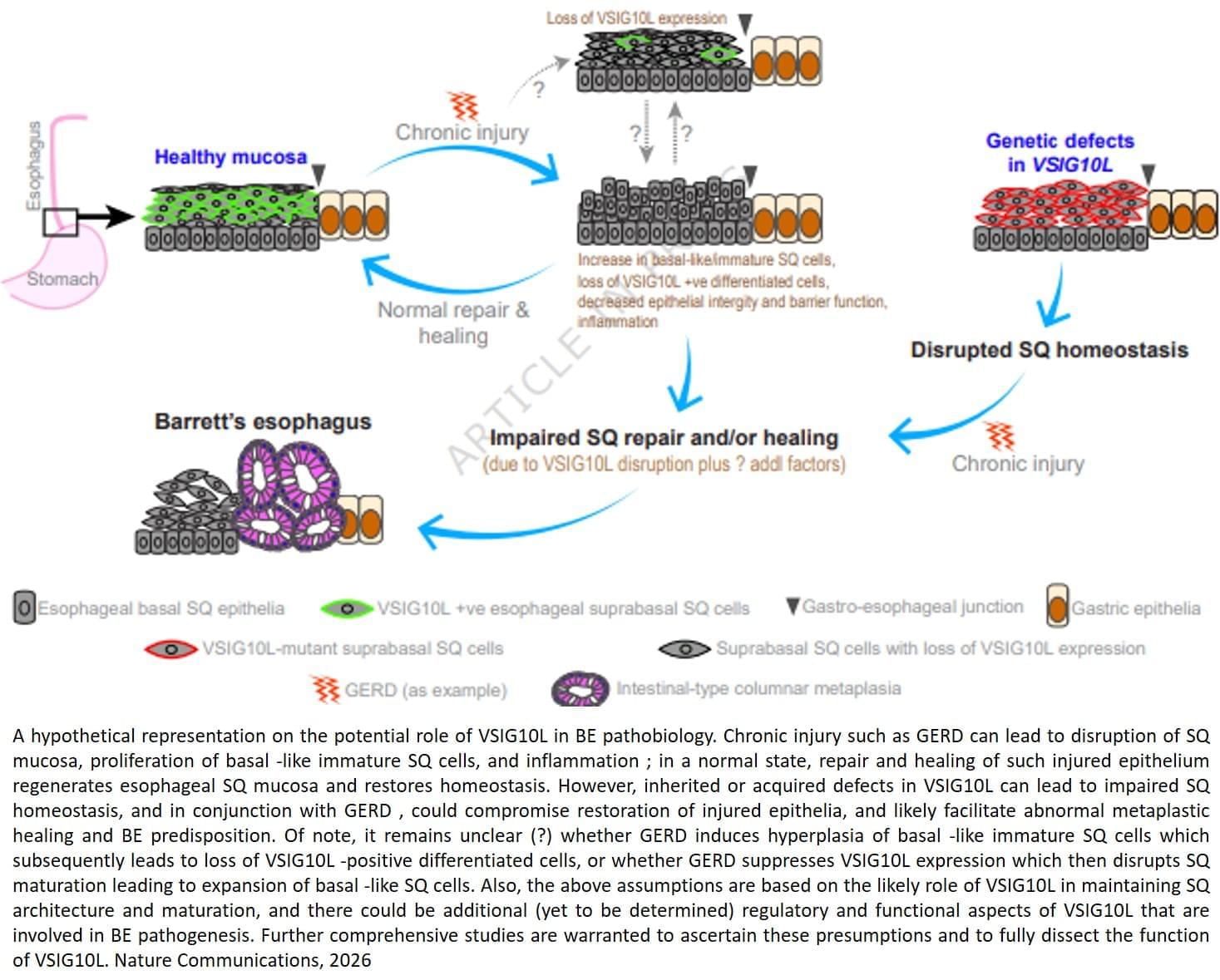
But the molecular factors responsible for the onset of Barrett’s esophagus remain poorly understood.
The findings, published in Nature Communications, combined family studies, laboratory experiments and genetically engineered mouse models to identify and understand how genetic defects contribute to disease development.
The team sequenced and analyzed genetic material of 684 people from 302 families where multiple members developed Barrett’s esophagus or esophageal cancer. They discovered that a subset of affected family members carry inherited mutations in a gene called VSIG10L.
“We found that this gene acts like a quality control system for the esophageal lining,” said the lead researcher. “When it’s defective, the cells do not mature properly and the protective barrier in the esophageal lining becomes weak, allowing stomach bile acid to cause tissue changes that enhances the risk of developing Barrett’s esophagus.”
When researchers genetically engineered mice with human-equivalent VSIG10L mutations, they found that the esophageal lining became disrupted structurally and molecularly, according to the author. The study found that when the mice were exposed to bile acid, they developed Barrett’s-like disease over time, effectively replicating the disease’s progression in humans.
These genetically engineered mice also represent the first animal model for Barrett’s esophagus based directly on human genetic predisposition to the disease, the author said.
With VSIG10L shown to be a key gene in maintaining esophageal health, family members can now be screened for genetic variants to identify those at a high-risk of developing Barrett’s esophagus or esophageal cancer. ScienceMission sciencenewshighlights.
TIL therapy for glioblastoma.
Tumor infiltrating lymphocyte (TIL) therapy has demonstrated encouraging efficacy in melanoma and nonsmall-cell lung cancer (NSCLC), and is now being explored for glioblastoma despite its immunologically ‘cold’ microenvironment.
Recent studies confirm that functional TILs can be expanded from cold tumors such as glioblastoma, including solid tumor resections and aspirates, overcoming previous feasibility concerns.
Advances in cytokine support, gene editing, and artificial antigen-presenting cells (APCs) are improving TIL persistence, cytotoxicity, and manufacturing scalability.
Focused ultrasound and nanoparticle delivery offer innovative solutions to enhance TIL infiltration across the blood– brain barrier. Integration of spatial multi-omics enables high-resolution mapping of immune niches and identification of tumorreactive clones.
Combination strategies with checkpoint blockade, myeloid modulation, and oncolytic virotherapy are emerging as rational paths to enhance TIL efficacy sciencenewshighlights ScienceMission https://sciencemission.com/TIL-therapy-17895
In All Tomorrows by C. M. Kosemen, also known as Nemo Ramjet, humanity’s distant descendants are reshaped across millions of years into wildly divergent “post-human” species after being genetically engineered by the godlike alien Qu. These forms range from tiny, almost vermin-like organisms and sessile, colony-bound beings to aquatic leviathans, aerial gliders, and towering, heavily built giants as each adapted to extreme planetary environments and radically different evolutionary pressures. Some retain echoes of recognizable humanity, while others are so transformed they blur the line between animal, ecosystem, and living architecture. In this size comparison, we’ll explore the full spectrum of these post-human forms, from the smallest engineered remnants to the most massive macro-organic descendants.
Credits:
https://all-tomorrows.fandom.com/wiki/Qu.
https://speculativeevolution.fandom.com/wiki/All_Tomorrows.
FAIR-USE COPYRIGHT DISCLAIMER Copyright Disclaimer under Section 107 of the Copyright Act 1976, allowance is made for “fair use” for purposes such as criticism, commenting, news reporting, teaching, scholarship, and research. Fair use is a use permitted by copyright statute that might otherwise be infringing. Non-profit, educational or personal use tips the balance in favour of fair use. Nutbug does not own the rights to these videos and pictures. They have, in accordance with fair use, been repurposed with the intent of educating and inspiring others. However, if any content owners would like their images removed, please contact us by email [email protected]

A new study led by the Institute for Bioengineering of Catalonia (IBEC) has unveiled the first biomaterial that is not only waterproof but actually becomes stronger in contact with water. The material is produced by the incorporation of nickel into the structure of chitosan, a chitinous polymer obtained from discarded shrimp shells. The development of this new biomaterial marks a departure from the plastic-age mindset of making materials that must isolate from their environment to perform well. Instead, it shows how sustainable materials can connect and leverage their environment, using their surrounding water to achieve mechanical performance that surpasses common plastics.
Plastics have become an integral part of modern society thanks to their durability and resistance to water. However, precisely these properties turn them into persistent disruptors of ecological cycles. As a result, unrecovered plastic is accumulating across ecosystems and becoming an increasingly ubiquitous component of global food chains, raising growing concerns about potential impacts on human health.
In an effort to address this challenge, the use of biomaterials as substitutes for conventional plastics has long been explored. However, their widespread adoption has been limited by a fundamental drawback: Most biological materials weaken when exposed to water. Traditionally, this vulnerability has forced engineers to rely on chemical modifications or protective coatings, thereby undermining the sustainability benefits of biomaterial-based solutions.
3D bioprinting, in which living tissues are printed with cells mixed into soft hydrogels, or “bio-inks,” is widely used in the field of bioengineering for modeling or replacing the tissues in our bodies. The print quality and reproducibility of tissues, however, can face challenges. One of the most significant challenges is created simply by gravity—cells naturally sink to the bottom of the bioink-extruding printer syringe because the cells are heavier than the hydrogel around them.
“This cell settling, which becomes worse during the long print sessions required to print large tissues, leads to clogged nozzles, uneven cell distribution, and inconsistencies between printed tissues,” explains Ritu Raman, the Eugene Bell Career Development Professor of Tissue Engineering and assistant professor of mechanical engineering at MIT.
“Existing solutions, such as manually stirring bioinks before loading them into the printer, or using passive mixers, cannot maintain uniformity once printing begins.”

G-protein-coupled receptors (GPCRs) are one of the largest families of cell surface proteins in the human body that recognize hormones, neurotransmitters, and drugs. These receptors regulate a wide range of physiological processes and are the targets of more than 30% of currently marketed drugs. The histamine H1 receptor (H1R) is one such GPCR subtype that plays a key role in mediating allergic reactions, inflammation, vascular permeability, airway constriction, wakefulness, and cognitive functions in the human body. While antihistamines primarily target H1R, current drugs can exhibit limited therapeutic efficacy, prompting researchers to look at H1R ligands from new perspectives.
Recently, the importance of drug design based not only on the affinity or binding energy between a compound and its target protein, but also on its components, enthalpy, and entropy, has been recognized as crucial for rational drug design. In particular, enthalpy–entropy compensation has emerged as a key concept for understanding ligand selectivity and isomer specificity. However, direct experimental measurement of these thermodynamic parameters has been limited to cell surface proteins, such as GPCRs.
Addressing this gap, a research team led by Professor Mitsunori Shiroishi from the Department of Life System Engineering, Tokyo University of Science (TUS), Japan, systematically investigated the binding thermodynamics of the H1R. The team included Mr. Hiroto Kaneko (first-year doctoral student) and Associate Professor Tadashi Ando from TUS, among others. Their study was published online in ACS Medicinal Chemistry Letters on January 26, 2026.
A team of researchers has biologically engineered T cells with currently available Alzheimer’s drugs in order to directly attack the characteristic amyloid plaques of Alzheimer’s disease.
Building on the current paradigm
Most Alzheimer’s treatments used in the clinic are-mabs, monoclonal antibodies that are designed to attack the amyloid beta plaques that accumulate in the brains of people with Alzheimer’s. However, while they have been found to have enough meaningful benefits in clinical trials to be approved by the FDA, they are not a cure, and some analyses question their effectiveness [1].

Researchers have developed a highly sensitive light-based sensor that can detect extremely low concentrations of cancer biomarkers in the blood. The new technology could one day make it possible to spot early signs of cancer and other conditions using a simple blood test.
Biomarkers such as proteins, DNA or other molecules can be used to reveal the presence, progression or risk of cancer and other diseases. However, one of the main challenges in early disease diagnosis is the extremely low concentration of biomarkers present at the onset.
“Our sensor combines nanostructures made of DNA with quantum dots and CRISPR gene editing technology to detect faint biomarker signals using a light-based approach known as second harmonic generation (SHG),” said research team leader Han Zhang from Shenzhen University in China.