Discover how Artificial Intelligence in Cancer Drug Discovery accelerates target identification, drug design, biomarkers, and clinical trials.
Category: biotech/medical – Page 90
Discovery of brain parasite’s unique control protein offers hope for better toxoplasmosis treatments
Rajshekhar Gaji was staring at something that should not exist. Under his microscope, parasites that should have been thriving were instead dying—completely unable to survive without a protein his lab had managed to switch off.
“It was an amazing day,” said Gaji, assistant professor of parasitology at the Virginia–Maryland College of Veterinary Medicine. That moment of discovery could eventually help the 40 million Americans walking around with a microscopic parasite permanently residing in their brains.
The findings are published in the journal mSphere.

Gene therapy delivers lasting immune protection in children with rare disorder
An experimental gene therapy developed by researchers at UCLA, University College London and Great Ormond Street Hospital has restored and maintained immune system function in 59 of 62 children born with ADA-SCID, a rare and deadly genetic immune disorder.
Severe combined immunodeficiency due to adenosine deaminase deficiency, or ADA-SCID, is caused by mutations in the ADA gene, which creates an enzyme essential for immune function. For children with the condition, day-to-day activities like going to school or playing with friends can lead to dangerous, life-threatening infections. If untreated, ADA-SCID can be fatal within the first two years of life.
The current standard treatments— bone marrow transplant from a matched donor or weekly enzyme injections—come with limitations and potential long-term risks.

New world record set for fastest human whole genome sequencing
Boston Children’s Hospital, along with Broad Clinical Labs and Roche Sequencing Solutions, has demonstrated that rapid genomic sequencing and interpretation are achievable in a matter of hours. This milestone not only sets a Guinness World Records for the fastest human whole genome sequencing to date but represents a significant clinical development that would expedite more precise treatments for critically ill babies in the NICU.
The team’s pilot data are published in the New England Journal of Medicine.
Current clinically available rapid genomic sequencing options take days (from sample receipt to report) at best, yet many critical care decisions in the NICU need to occur within a matter of hours. While there have been prior demonstrations of genome-sequencing within hours, none up to now have been scalable or feasible for routine use.
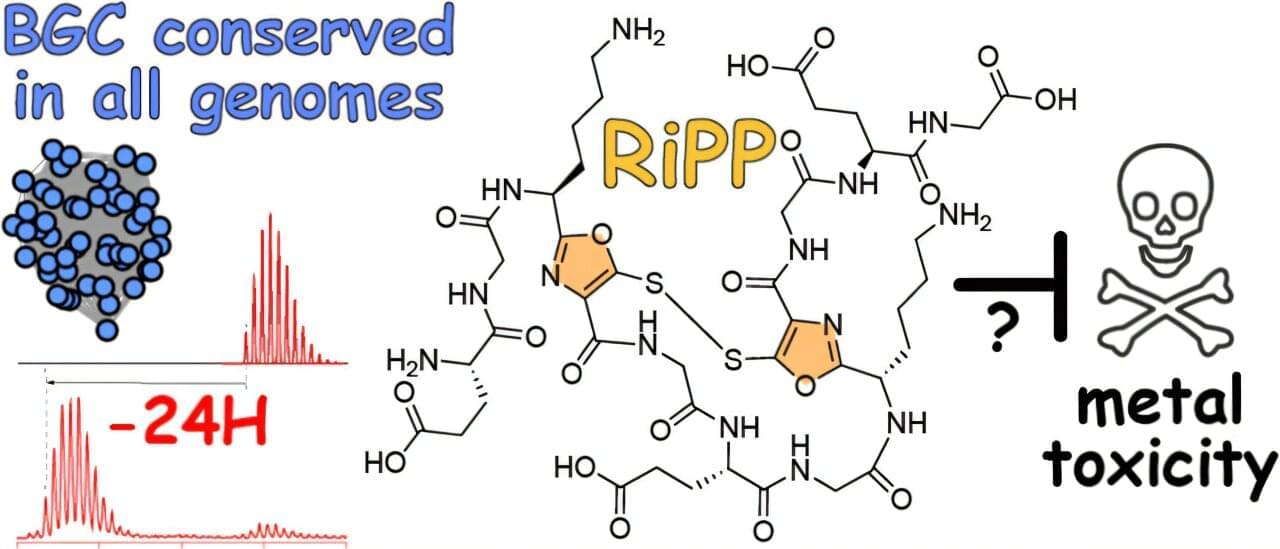
Undergrads uncover conserved copper-binding gene cluster in marine bacteria
This fall, 20 Georgia Tech students published a paper—the culmination of work done during a semester-long laboratory course. During the semester, students analyzed genomes sequenced from marine samples collected in Key West, Florida—doing hands-on original bioinformatics research on par with graduate students and working with bioinformatics tools to explore drug discovery potential.
The course, BIOS 4,590, is a research project lab for senior biology majors that provides an opportunity for professors to share their expertise with students in a hands-on environment. In his class, Associate Professor Vinayak (Vinny) Agarwal, who holds joint appointments in the School of Chemistry and Biochemistry and School of Biological Sciences, aimed to introduce undergraduates to advanced bioinformatics tools through applied research using new-to-science raw data.
The resulting paper, “Phylogenomic Identification of a Highly Conserved Copper-Binding RiPP Biosynthetic Gene Cluster in Marine Microbulbifer Bacteria,” which was recently published in ACS Chemical Biology, involves the historically understudied genus of Microbulbifer, a type of bacteria often associated with sponges and corals. These microbial communities are rich sources of natural products, small biological molecules often associated with medicine and drug discovery.
Atom-scale stencil patterns help nanoparticles take new shapes and learn new tricks
Inspired by an artist’s stencils, researchers have developed atomic-level precision patterning on nanoparticle surfaces, allowing them to “paint” gold nanoparticles with polymers to give them an array of new shapes and functions.
The “patchy nanoparticles” developed by University of Illinois Urbana-Champaign researchers and collaborators at the University of Michigan and Penn State University can be made in large batches, used for a variety of electronic, optical or biomedical applications, or used as building blocks for new complex materials and metamaterials.
Led by Qian Chen, an Illinois professor of materials science and engineering, the researchers report their findings in the journal Nature.

Why some quantum materials stall while others scale
People tend to think of quantum materials—whose properties arise from quantum mechanical effects—as exotic curiosities. But some quantum materials have become a ubiquitous part of our computer hard drives, TV screens, and medical devices. Still, the vast majority of quantum materials never accomplish much outside of the lab.
What makes certain quantum materials commercial successes and others commercially irrelevant? If researchers knew, they could direct their efforts toward more promising materials—a big deal since they may spend years studying a single material.
Now, MIT researchers have developed a system for evaluating the scale-up potential of quantum materials. Their framework combines a material’s quantum behavior with its cost, supply chain resilience, environmental footprint, and other factors.

Our team of physicists inadvertently generated the shortest X-ray pulses ever observed
X-ray beams aren’t used just by doctors to see inside your body and tell whether you have a broken bone. More powerful beams made up of very short flashes of X-rays can help scientists peer into the structure of individual atoms and molecules and differentiate types of elements.
But getting an X-ray laser beam that delivers super short flashes to capture the fastest processes in nature isn’t easy—it’s a whole science in itself.
Radio waves, microwaves, the visible light you can see, ultraviolet light and X-rays are all exactly the same phenomenon: electromagnetic waves of energy moving through space. What differentiates them is their wavelength. Waves in the X-ray range have short wavelengths, while radio waves and microwaves are much longer. Different wavelengths of light are useful for different things—X-rays help doctors take snapshots of your body, while microwaves can heat up your lunch.

DNA signaling cascades offer a better way to monitor drug therapy at home
Chemists at Université de Montréal have developed “signaling cascades” made with DNA molecules to report and quantify the concentration of various molecules in a drop of blood, all within five minutes.
Their findings, validated by experiments on mice, are published in the Journal of the American Chemical Society, and may aid efforts to build point-of-care devices for monitoring and optimizing the treatment of various diseases.
This result was achieved by a research group led by UdeM chemistry professor Alexis Vallée-Bélisle.
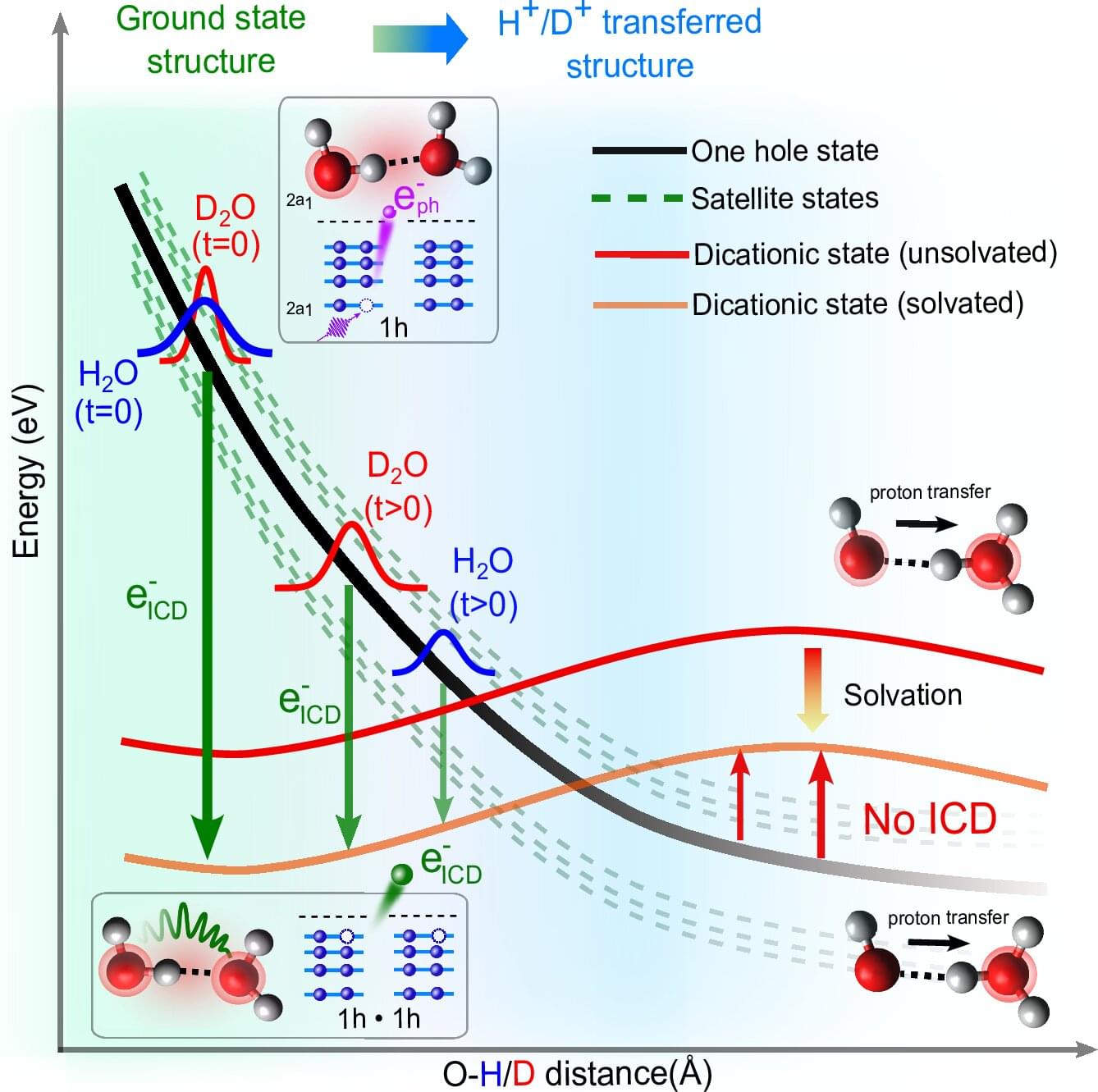
Generation of harmful slow electrons in water is a race between intermolecular energy decay and proton transfer
When high-energy radiation interacts with water in living organisms, it generates particles and slow-moving electrons that can subsequently damage critical molecules like DNA. Now, Professor Petr Slavíček and his bachelor’s student Jakub Dubský from UCT Prague (University of Chemistry and Technology, Prague) have described in detail one of the key mechanisms for the creation of these slow electrons in water, a process known as Intermolecular Coulombic Decay (ICD). Their powerful mathematical model successfully explains all the data from complex laser experiments conducted at ETH Zurich (Hans-Jakob Woerner team).
The work, which deepens the fundamental understanding of radiation chemistry, has been published in the journal Nature Communications.
A detailed knowledge of the processes in aqueous solutions, combined with advances in research technologies using high-energy radiation, is transforming the field of radiation chemistry. In the future, these insights could lead to significant changes in various fields, including medicine, particularly in developing more sensitive and controllable applications for devices based on ionizing radiation.