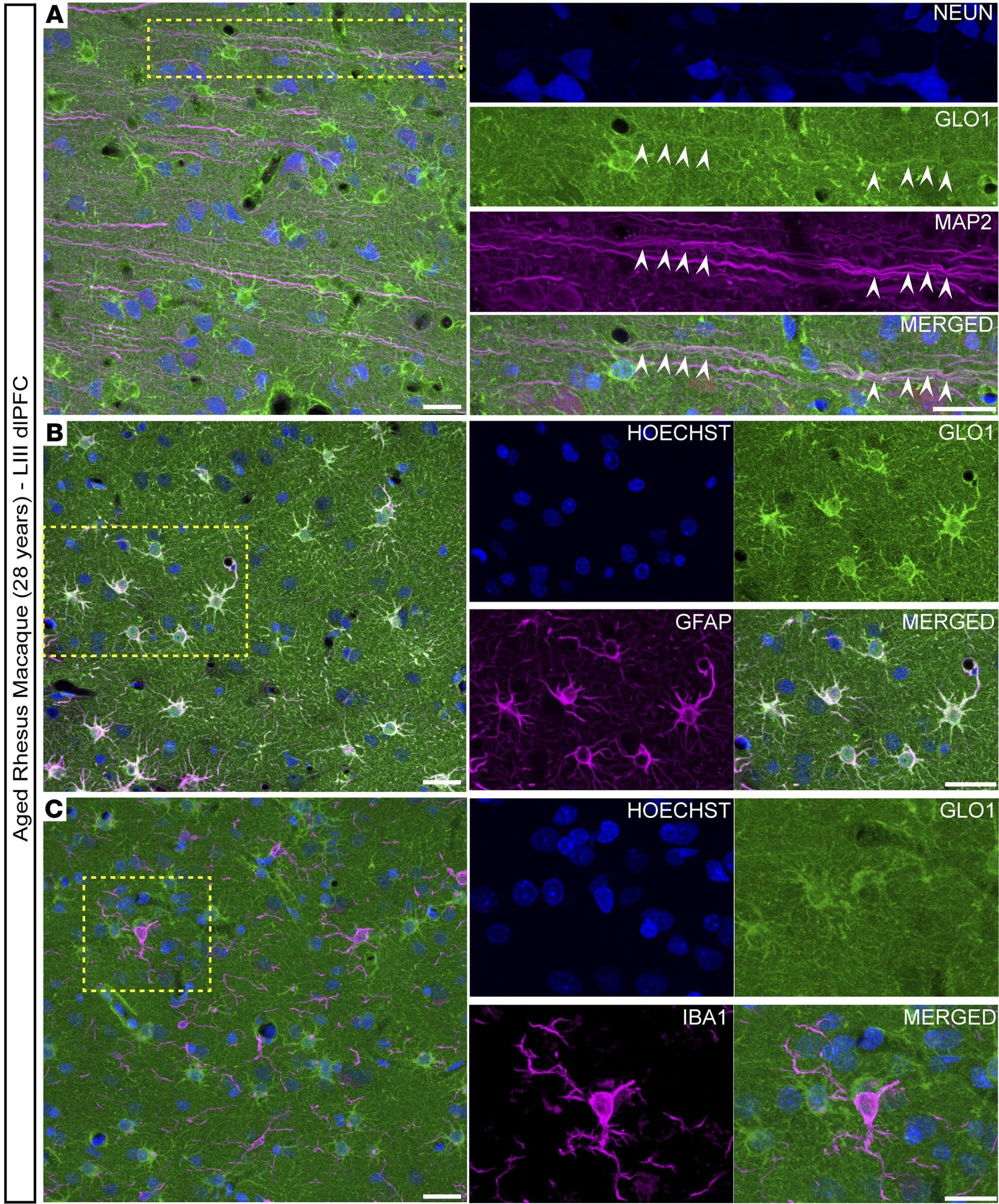
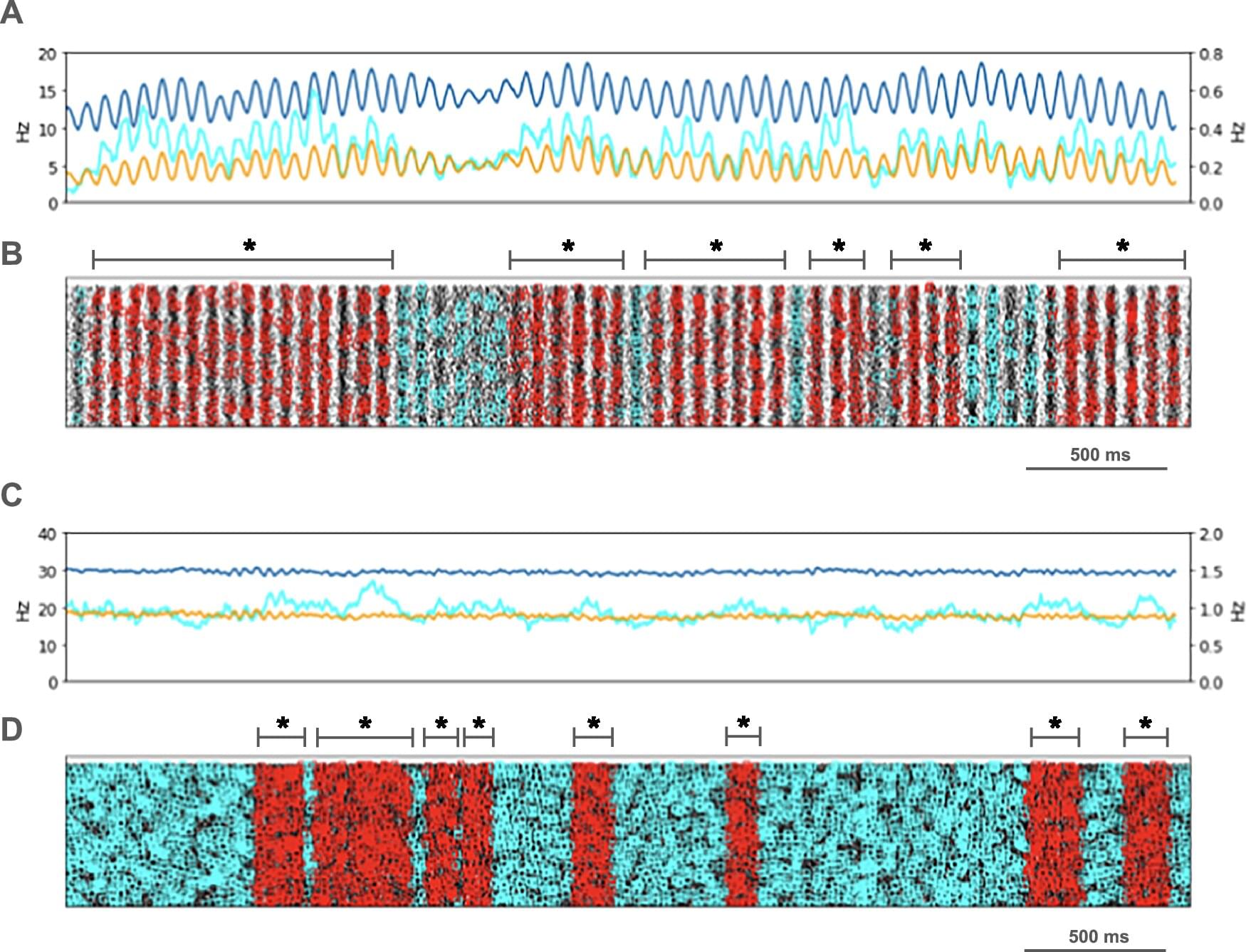
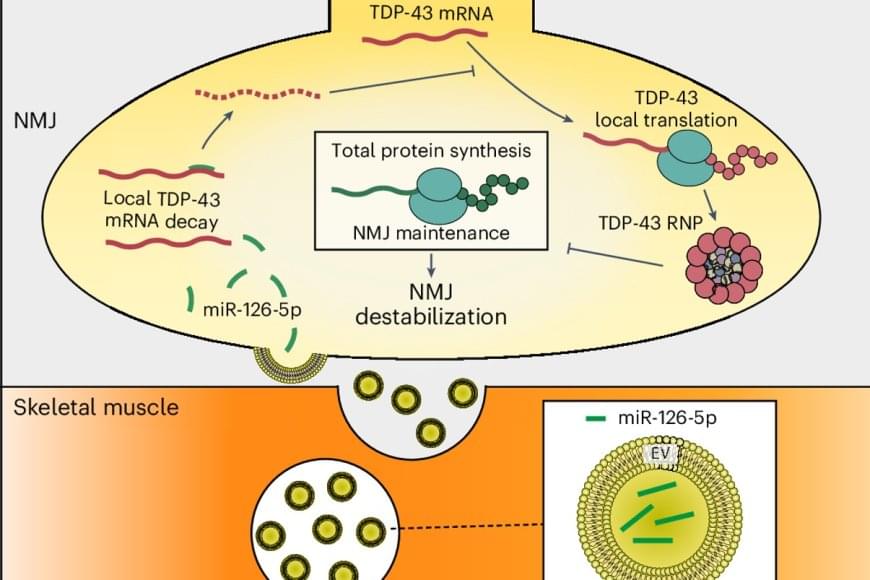
The living cell, once thought to be a precise molecular factory, is turning out to be more like an improvising jazz ensemble. The old dogma—one gene, one protein, one function—has collapsed. Molecular biologist Ewa Grzybowska argues that recent discoveries show that proteins can switch folds, shift shapes, or even remain gloriously unstructured, improvising their roles as they go. Genes are not blueprints but texts, open to continuous interpretation by cells. Life, it turns out, is not built like a machine but is instead fluid, improvisational, and brimming with creative possibility.
1. The old paradigms in biology
When Watson and Crick decoded DNA in 1953 and the mechanism of protein-making was discovered, we obtained an extraordinary tool to explain the inner workings of life. The basic principle of making one protein from one DNA template (gene) with the assistance of one messenger RNA (mRNA) and several well-defined amino-acid-transporting RNAs (tRNAs) was so successful that it has been enshrined in millions of textbooks and not questioned for a long time.