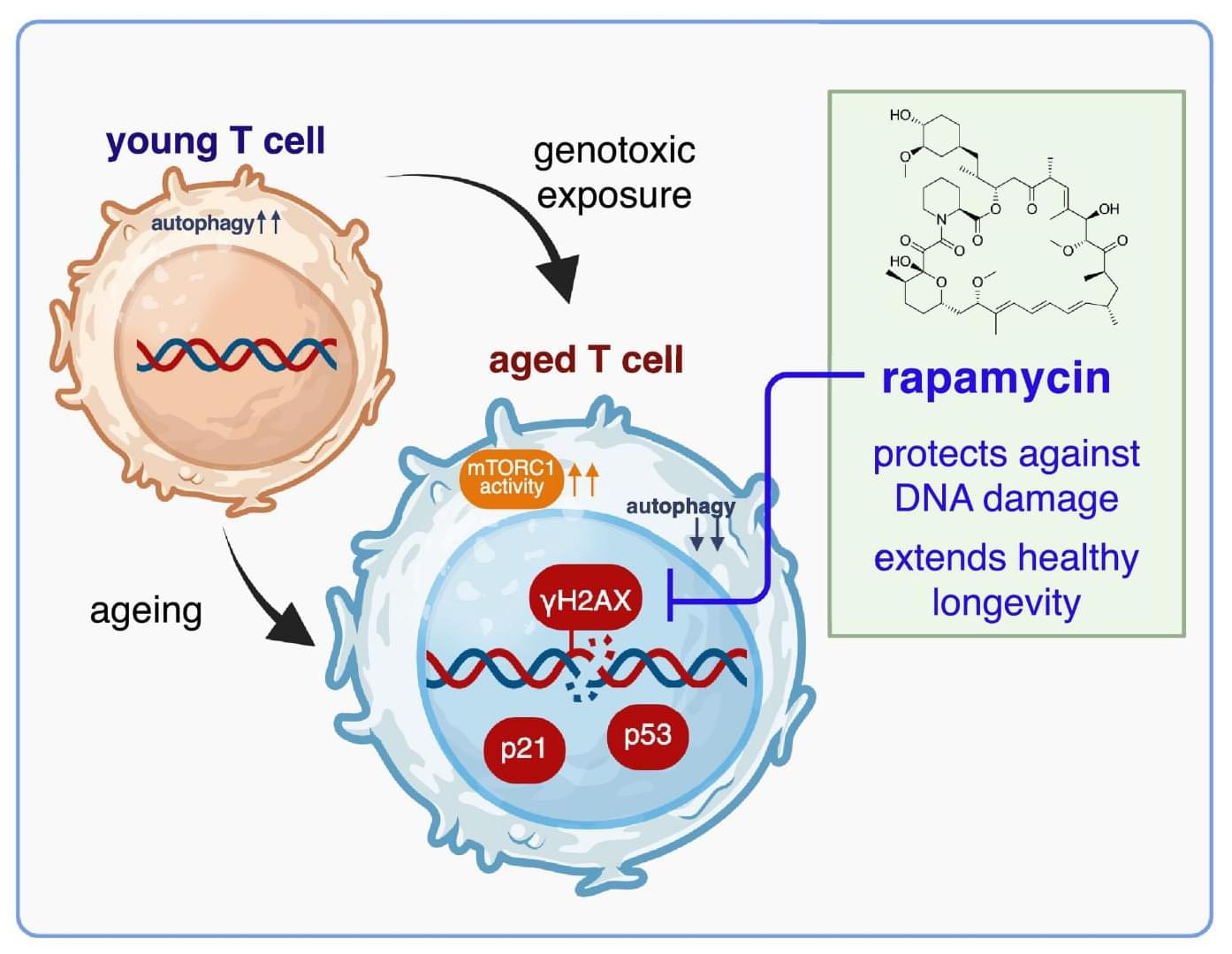
Severe blood loss remains the primary cause of death from combat injuries. To address this challenge, a research team at KAIST that included an active duty Army Major set out to develop a faster and more reliable way to stop bleeding.
Their work led to a next-generation powder-type hemostatic agent that can halt bleeding within one second when sprayed directly onto a wound, offering a potential breakthrough for saving lives on the battlefield.