The Gallery of Fluid Dynamics winners’ circle includes drops shaken loose from a wire, odor diffusion in a breeze, and droplets acting quantum-like.


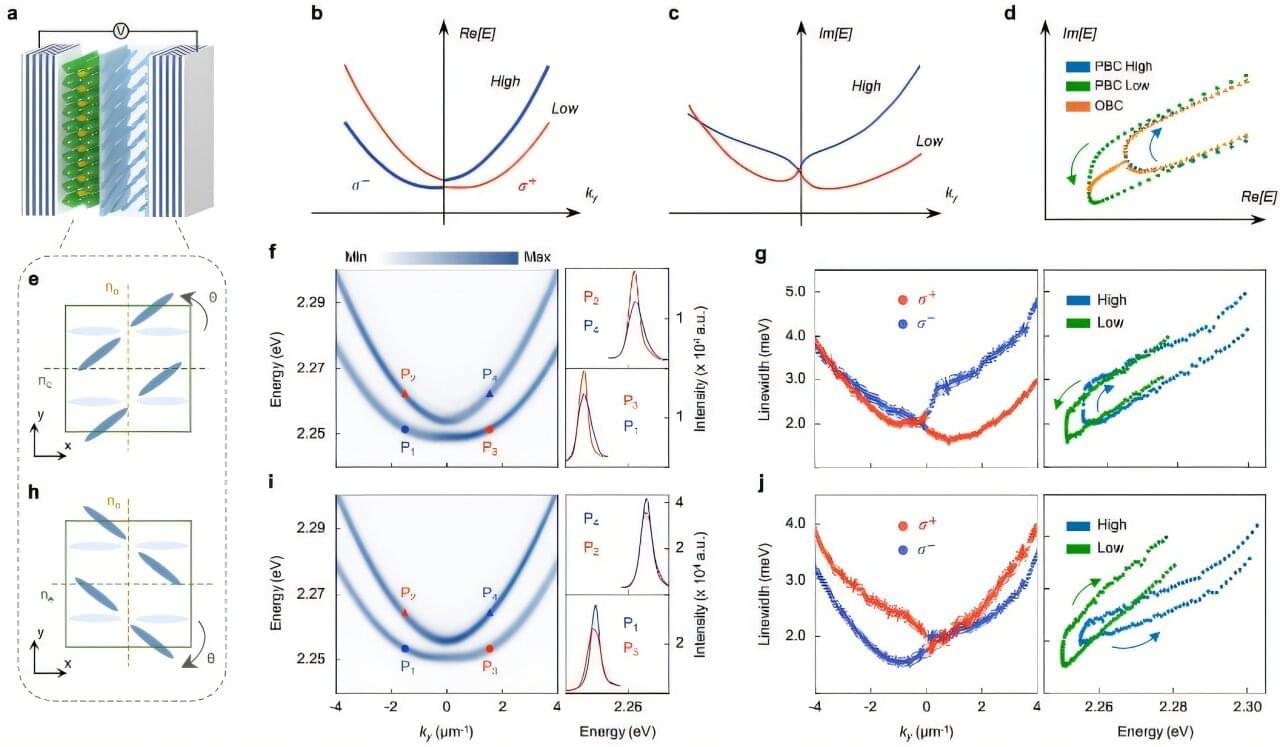
Properties that remain unchanged when materials are stretched or bent, which are broadly referred to as topological properties, can contribute to the emergence of unusual physical effects in specific systems.
Over the past few years, many physicists have been investigating the physical effects emerging from the topology of non-Hermitian systems, open systems that exchange energy with their surroundings.
Researchers at Nanyang Technological University and the Australian National University set out to probe non-Hermitian topological phenomena in systems comprised of light and matter particles that strongly interact with each other.

Many researchers have spent decades attempting to decode biblical descriptions and link them to verifiable historical events. One such description is that of the Star of Bethlehem—a bright astronomical body that was said to lead the Magi to Jesus shortly after his birth.
Although many attempts have been made to link the Star of Bethlehem to astronomical bodies, the unique motion of the “star” did not quite fit any known object. However, a new research study, published in Journal of the British Astronomical Association, describes a likely candidate for the bright object seen above Bethlehem over 2000 years ago—a comet described in an ancient Chinese text.
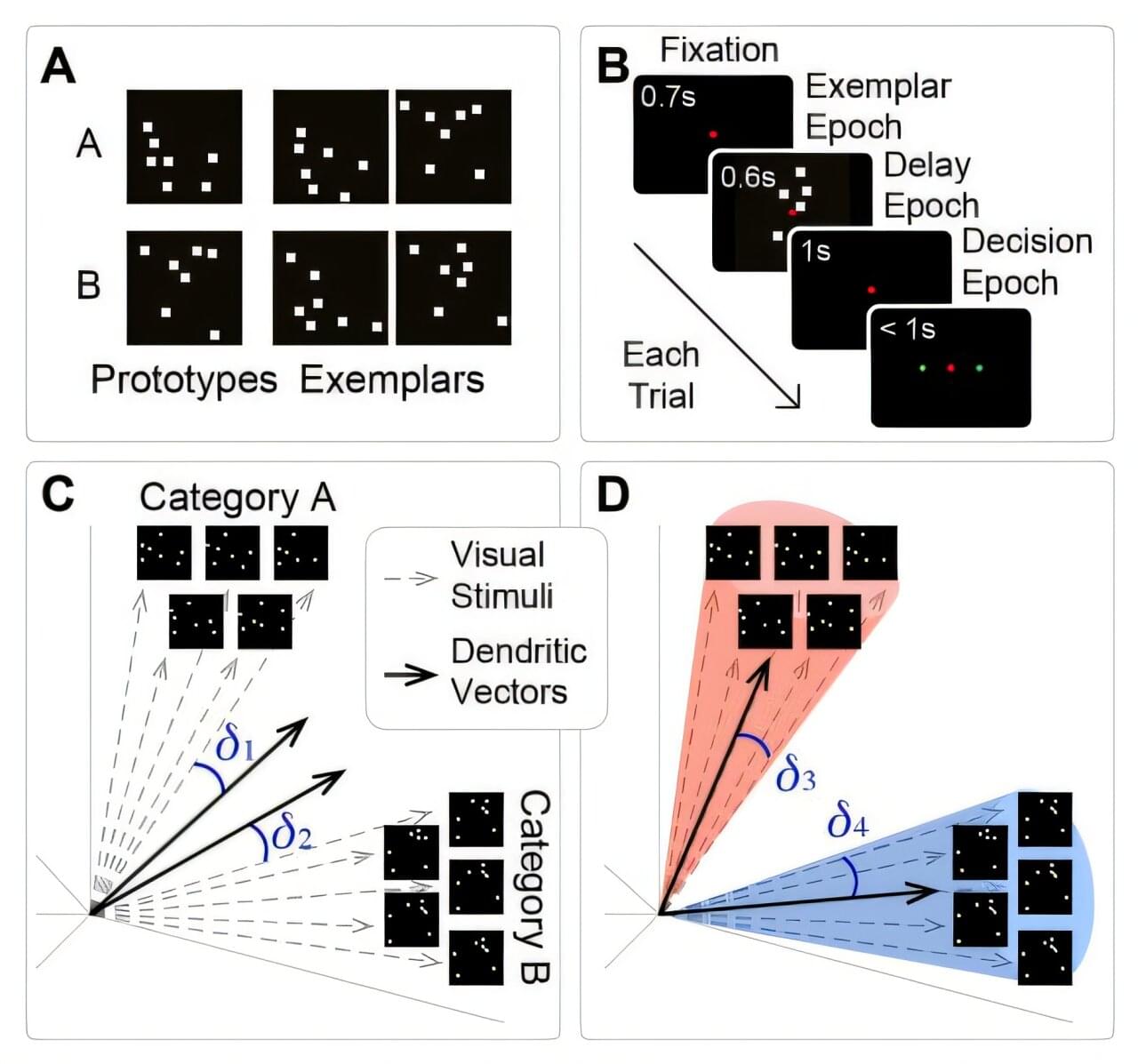
A new computational model of the brain based closely on its biology and physiology has not only learned a simple visual category learning task exactly as well as lab animals, but even enabled the discovery of counterintuitive activity by a group of neurons that researchers working with animals to perform the same task had not noticed in their data before, reports a team of scientists at Dartmouth College, MIT, and the State University of New York at Stony Brook.


Astronomers have employed the Very Energetic Radiation Imaging Telescope Array System (VERITAS) to observe a mysterious gamma-ray emitting source designated HESS J1857+026. Results of the observational campaign, published December 19 on the pre-print server arXiv, shed more light on the nature of this source.

When you splurge on a cocktail in a bar, the drink often comes with a slab of aesthetically pleasing, perfectly clear ice. The stuff looks much fancier than the slightly cloudy ice you get from your home freezer. How do they do this?
Clear ice is actually made from regular water—what’s different is the freezing process.
With a little help from science, you can make clear ice at home, and it’s not even that tricky. However, there are quite a few hacks on the internet that won’t work. Let’s dive into the physics and chemistry involved.
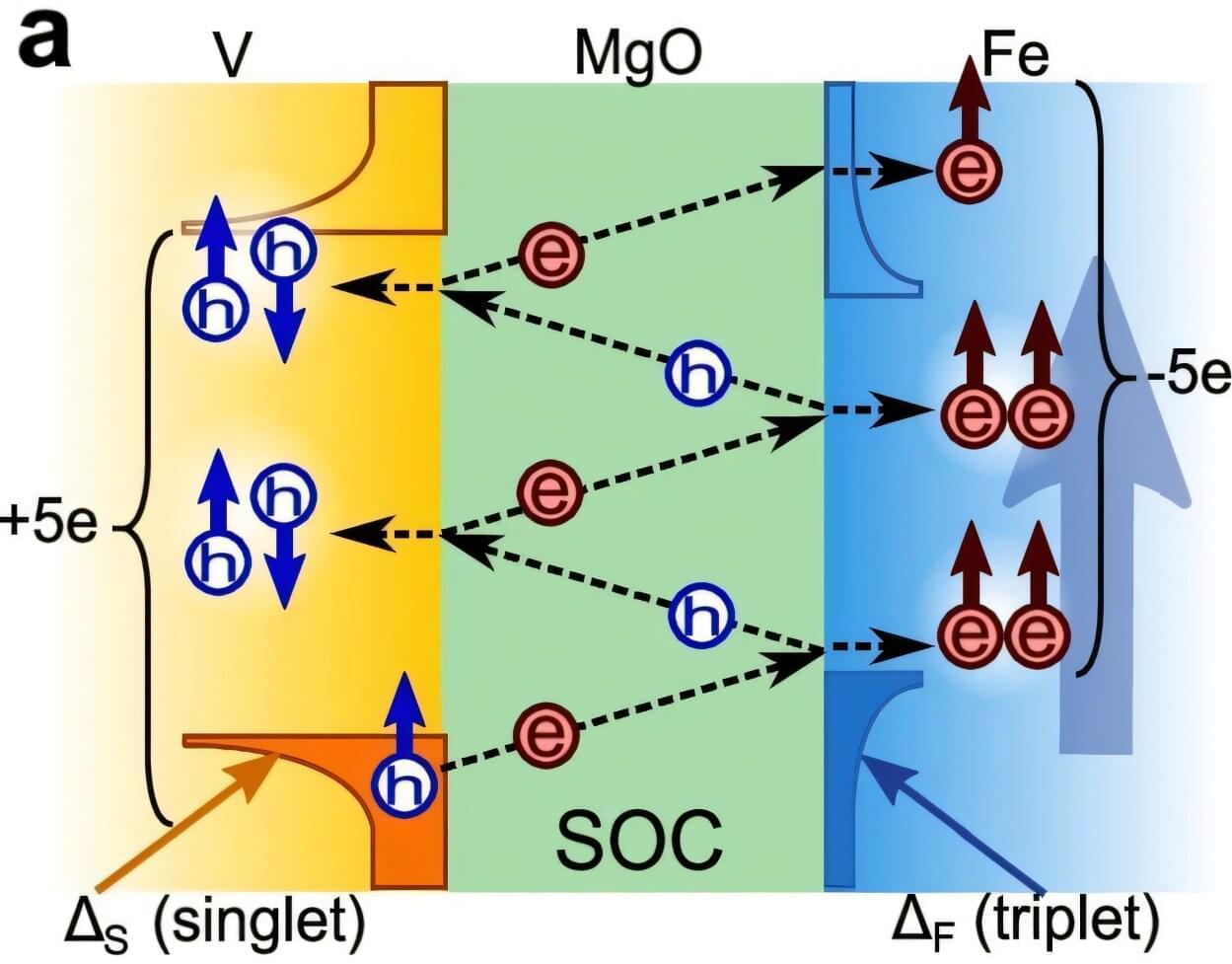
Separate two superconductors with a thin layer of material and something strange happens.
Their superconductivity—a property driven by paired electrons that allows electricity to flow without energy loss—can leak into the barrier and link together, synchronizing their behavior despite the separation.
This device is known as a Josephson junction. It’s the foundational building block of quantum computers and advances of it won the 2025 Nobel Prize in Physics.

These days, large language models can handle increasingly complex tasks, writing complex code and engaging in sophisticated reasoning. But when it comes to four-digit multiplication, a task taught in elementary school, even state-of-the-art systems fail. Why?
A new paper posted to the arXiv preprint server by University of Chicago computer science Ph.D. student Xiaoyan Bai and faculty co-director of the Data Science Institute’s Novel Intelligence Research Initiative Chenhao Tan finds answers by reverse-engineering failure and success.
They worked with collaborators from MIT, Harvard University, University of Waterloo and Google DeepMind to probe AI’s “jagged frontier”—a term for its capacity to excel at complex reasoning yet stumble on seemingly simple tasks.
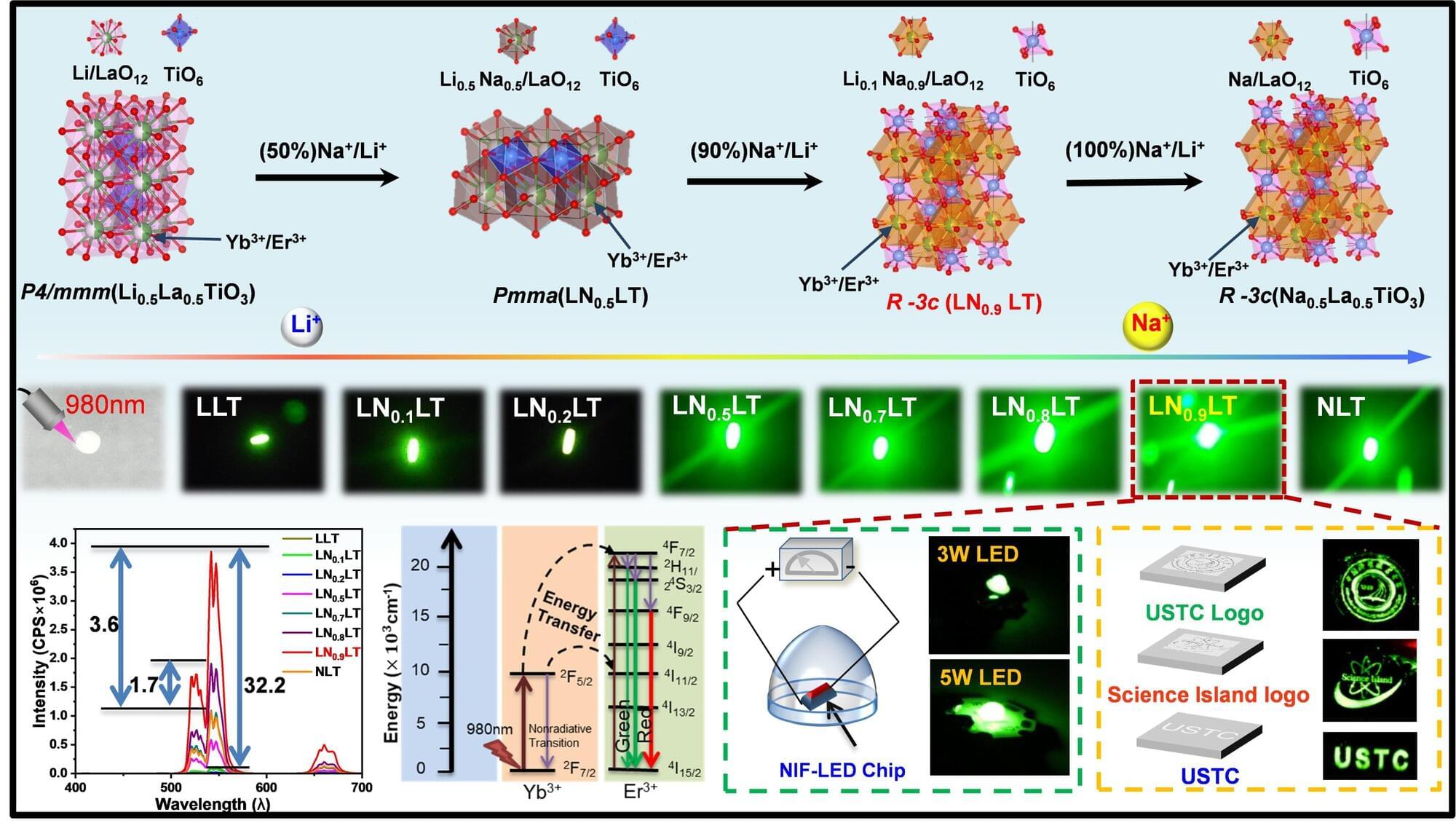
Researchers at the Hefei Institutes of Physical Science of the Chinese Academy of Sciences have developed a new way to significantly enhance upconversion luminescence in oxide perovskites, a class of materials known for their thermal and chemical stability but limited optical efficiency.
Led by Professor Jiang Changlong, the team introduced a dual-cation substitution strategy in titanate perovskites by precisely adjusting the sodium-to-lithium ratio at the crystal’s A-site. This controlled substitution triggers a structural transition that improves energy transfer between rare-earth ions, resulting in a marked increase in luminescence intensity and quantum yield.
The findings are published in Journal of Alloys and Compounds.