In the pursuit of sustainable energy solutions, the quest for more efficient solar cells is paramount. Organic photovoltaic cells have emerged as a promising alternative to traditional silicon-based counterparts due to their flexibility and cost-effectiveness. However, optimizing their performance remains a significant challenge.
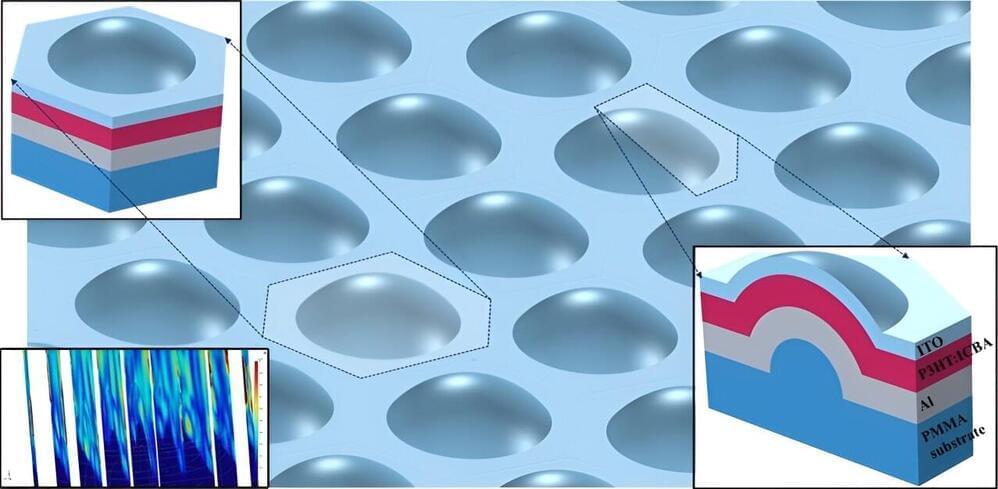
In a pioneering move, new research from Abdullah Gül University (Türkiye) reimagines the structure of organic photovoltaic cells, opting for a hemispherical shell shape to unlock unprecedented potential in light absorption and angular coverage.
As reported in the Journal of Photonics for Energy, this innovative configuration aims to maximize light absorption and angular coverage, promising to redefine the landscape of renewable energy technologies. The study presents advanced computational analysis and comparative benchmarks to spotlight the remarkable capabilities of this new design.





 עברית (Hebrew)
עברית (Hebrew)