I drive my car regularly between my home and lab. But one day, deep in my thoughts while driving, I suddenly found a speed breaker had cropped up overnight on

In a historic first for the laboratory, CERN has received $1 billion in private donations to support the development of the Future Circular Collider (FCC).
This philanthropic backing marks a shift in CERN’s 72-year funding history as it seeks to bridge the gap for the project’s estimated $18 billion price tag.
It comes from the Breakthrough Prize Foundation, the Eric and Wendy Schmidt Fund, and billionaire entrepreneurs John Elkann and Xavier Niel. Together, they pledged a combined $1 billion in late December 2025 to jumpstart the project.
In this report, researchers link thermogenic adipose tissue (brown/beige fat), best known for heat production, to blood-pressure control via direct fat–blood vessel communication. Using mouse models engineered to lose beige fat identity (via adipocyte-specific disruption of PRDM16), they observed elevated arterial pressure alongside perivascular remodeling, including fibrotic tissue accumulation and marked vascular hypersensitivity to the vasoconstrictor hormone angiotensin II. Mechanistically, loss of beige fat identity increased secretion of QSOX1 (quiescin sulfhydryl oxidase 1), which activated pro-fibrotic gene programs in vascular cells and promoted vessel stiffening; blocking this pathway (including genetic removal of QSOX1 in the model) restored healthier vascular signaling and function. The authors characterize this as a previously underappreciated, obesity-independent axis by which the “quality” (thermogenic vs white-like) of perivascular fat influences vascular stiffness and responsiveness to pressor signals, suggesting QSOX1 and related adipose-derived signals as potential precision targets for future antihypertensive therapies.
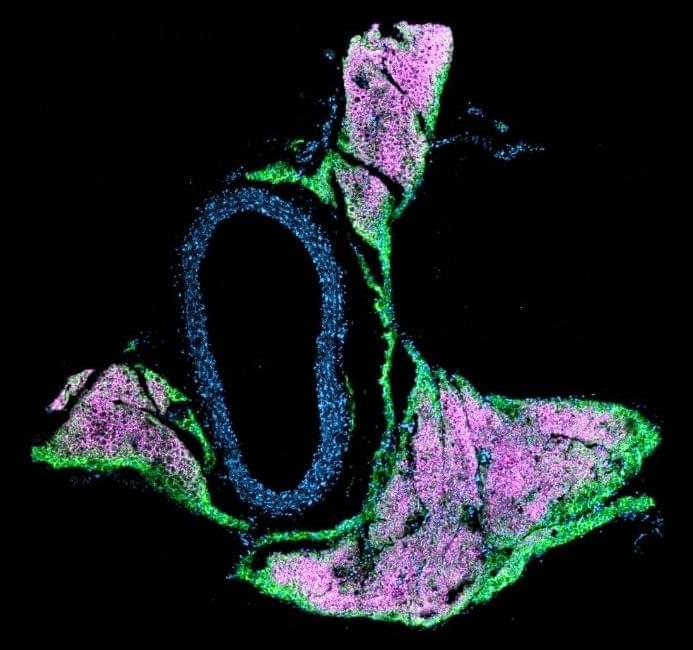
A mouse aorta with immunofluorescent tagging, emphasizing the close connection between vasculature and fat. (Credit: Cohen lab)
Obesity causes hypertension. Hypertension causes cardiovascular disease. And cardiovascular disease is the leading cause of death worldwide. While the link between fat and high blood pressure is clearly central to this deadly chain, its biological basis long remained unclear. What is it about fat that impacts vascular function and blood pressure control?
Now, a new study demonstrates how thermogenic beige fat—a type of adipose tissue, distinct from white fat, that helps the body burn energy—directly shapes blood pressure control. Building on clinical evidence that people with brown fat have lower odds of hypertension, the researchers created mouse models that cannot form beige fat (the thermogenic fat depot in mice that most closely resembles adult human brown fat) to watch what happens when this tissue is lost. They found that the loss of beige fat increases the sensitivity of blood vessels to one of the most important vasoconstricting hormones (angiotensin II)—and that blocking an enzyme involved in stiffening vessels and disrupting normal signaling can restore healthy vascular function in mice. These results, published in Science (opens in new window), reveal a previously unknown mechanism driving high blood pressure and point toward more precise therapies that target communication between fat and blood vessels.
Ray, you’ve made two predictions that I think are important. The first one, as you said, was the one you announced back in 1989: that we would reach human-level AI by 2029. And as you said, people laughed at it.
But there’s another prediction you’ve made: that we will reach the Singularity by 2045. There’s a lot of confusion here. In other words, if we reach human-level AI by 2029 and it then grows exponentially, why do we have to wait until 2045 for the Singularity? Could you explain the difference between these two?
It’s because that’s the point at which our intelligence will become a thousand times greater. One of the ways my view differs from others is that I don’t see it as us having our own intelligence—that is, biological intelligence—while AI exists somewhere else, and we interact with it by comparing human intelligence to AI.
Founder of XPRIZE and pioneer in exponential technologies. Building a world of Abundance through innovation, longevity, and breakthrough ventures.



“A strong magnetic field is very important for life on a planet,” said Dr. Miki Nakajima.
How can magnetic fields help determine the habitability of exoplanets? This is what a recent study published in Nature Astronomy hopes to address as a team of researchers from the University of Rochester and the University of California, Los Angeles investigated the formation processes that create magnetic fields on Earth and exoplanets slightly larger than Earth called super-Earths. This study has the potential to help scientists better understand planetary formation processes and the planetary conditions to search for life as we know it.
For the study, the researchers used a combination of laboratory experiments and computer models to simulate the formation processes of exoplanets, specifically focusing on the formation of the interior magma ocean responsible for generating the planet’s magnetic field like on Earth. The goal of the study was to estimate the long-term evolution of super-Earths, which are estimated to be between 1–10 Earth masses and 2–3 Earth radii. In the end, the researchers found that super-Earths between 3–6 Earth masses can produce magnetic fields that are stronger than Earths for up to several billion years.
“A strong magnetic field is very important for life on a planet,” said Dr. Miki Nakajima, who is an associate professor of Earth and Environmental Sciences at the University of Rochester and lead author of the study. “But most of the terrestrial planets in the solar system, such as Venus and Mars, do not have them because their cores don’t have the right physical conditions to generate a magnetic field. However, super-earths can produce dynamos in their core and/or magma, which can increase their planetary habitability.”
The media and tech landscape is undergoing a significant transformation driven by advancements in AI, technology, and new structures, enabling entrepreneurs and companies to achieve exponential growth and innovation ## ## Questions to inspire discussion.
Building Your Own Platform.
🚀 Q: How can writers escape traditional media constraints? A: Launch on decentralized platforms like Substack where you build your own brand and business as a “non-fungible writer”, potentially creating organizations 10x larger than traditional media companies you’d work for.
💰 Q: What makes writer-led platforms attractive investments? A: Platforms become cornerstone franchises when writers only succeed by making the platform successful, creating aligned incentives that generate significant returns while enabling top talent to build independent businesses.
📊 Q: What content opportunity exists in decentralized media? A: A barbell market is emerging with mainstream filler content on one end and massive untapped demand for high-quality niche content on the other, creating opportunities across various specialized domains.
Leveraging AI for Business.
Tesla is ending the one-time purchase option for Full Self-Driving (FSD) and shifting to a monthly subscription model, likely to recapture the value of the technology as it advances towards full autonomy and potential expansion into a robo-taxi fleet ##
## Questions to inspire discussion.
Investment Signal.
🎯 Q: Why is Tesla ending FSD one-time purchases after February 14?
A: Tesla is stopping FSD sales because autonomy is approaching a major inflection point where value will step-change when drivers are out of the loop, and Tesla wants to avoid locking in one-time payments at legacy prices before entering the real robo-taxi world.
Revenue Model Transformation.