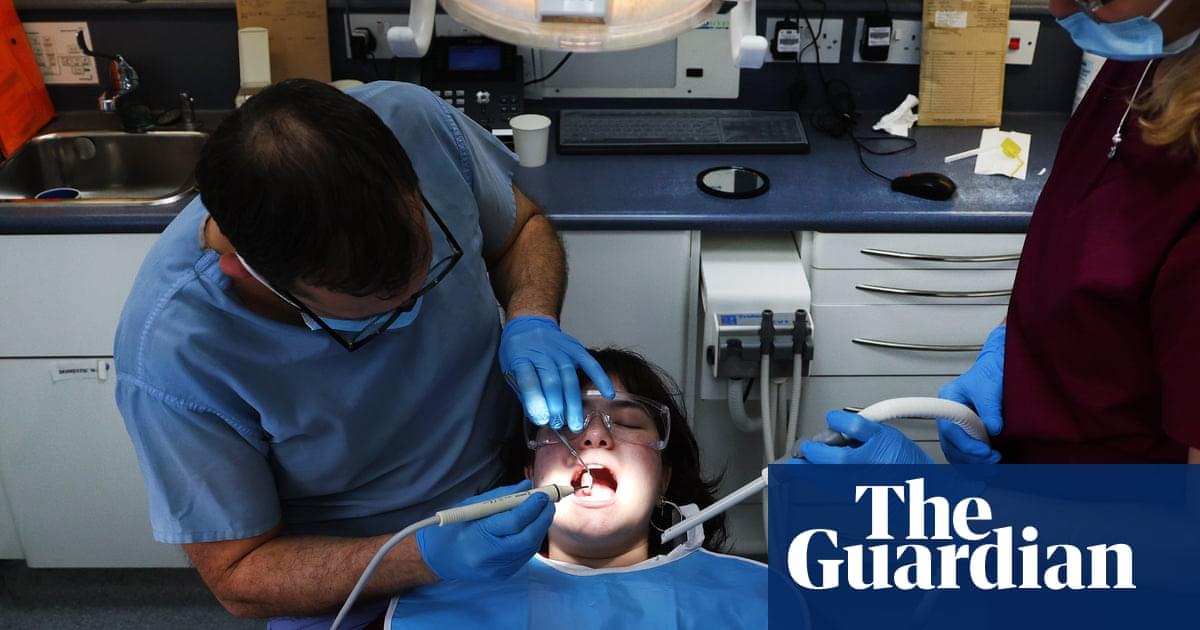
“Our oral health is connected to our general health,” said Dr Sadia Niazi, a senior clinical lecturer in endodontology at King’s College London. “We should never look at our teeth or dental disease as a separate entity.”
Root canal treatment is one of the most common – and perhaps most feared – dental procedures, though much of the anxiety derives from myths and misconceptions that hark back to the days of poor anaesthetics. The treatment is performed to treat infection or damage to the tooth’s pulp, the soft inner tissue of a tooth that contains nerves, blood vessels and connective tissue.
Read More
If a looming root canal treatment is putting a dampener on the week, take heart: having the procedure can drive health benefits that are felt throughout the body, according to research.
Patients who were successfully treated for root canal infections saw their blood sugar levels fall significantly over two years, suggesting that ridding the body of the problematic bacteria could help protect against type 2 diabetes.
Dentists also saw improvements in patients’ blood cholesterol and fatty acid levels, both of which are associated with heart health. Yet more benefits were seen around inflammation, a driver for cardiovascular disease and other chronic conditions.