Beef cattle genetically altered to be less susceptible to heat stress have been cleared for human consumption by the FDA.
Category: genetics – Page 323

Genetically modified pig hearts transplanted into two more patients
The team of researchers who transplanted a genetically modified pig’s heart into a living human earlier this year have completed two more pig heart transplant surgeries, setting the protocol for such operations.
In January this year, 57-year-old David Bennett became the first man on the planet to receive a heart from a genetically modified pig. Before this, researchers transplanted kidneys from similarly modified pigs into patients that were brain dead.
The organs are sourced from a company called Revivicor which uses genetic engineering to remove specific genes in the pigs to help in reducing transplant rejection while adding some that make the organs more compatible with the human immune system.

Global team of scientists discover new gene causing severe neurodevelopmental delays
The Neuro-Network.
𝐆𝐥𝐨𝐛𝐚𝐥 𝐭𝐞𝐚𝐦 𝐨𝐟 𝐬𝐜𝐢𝐞𝐧𝐭𝐢𝐬𝐭𝐬 𝐝𝐢𝐬𝐜𝐨𝐯𝐞𝐫 𝐧𝐞𝐰 𝐠𝐞𝐧𝐞 𝐜𝐚𝐮𝐬𝐢𝐧𝐠 𝐬𝐞𝐯𝐞𝐫𝐞 𝐧𝐞𝐮𝐫𝐨𝐝𝐞𝐯𝐞𝐥𝐨𝐩𝐦𝐞𝐧𝐭𝐚𝐥 𝐝𝐞𝐥𝐚𝐲𝐬
𝘼𝙣 𝙞𝙣𝙩𝙚𝙧𝙣𝙖𝙩𝙞𝙤𝙣𝙖𝙡 𝙩𝙚𝙖𝙢 𝙤𝙛 𝙧𝙚𝙨𝙚𝙖𝙧𝙘𝙝𝙚𝙧𝙨 𝙡𝙚𝙙 𝙗𝙮 𝙐𝘾 𝘿𝙖𝙫𝙞𝙨 𝙜𝙚𝙣𝙚𝙩𝙞𝙘𝙞𝙨𝙩 𝙎𝙪𝙢𝙖 𝙎𝙝𝙖𝙣𝙠𝙖… See more.
An international team of researchers led by UC Davis geneticist Suma Shankar has discovered a new gene implicated in a neurodevelopmental condition called DPH5-related diphthamide-deficiency syndrome. The syndrome is caused by DPH5 gene variants that may lead to embryonic death or profound neurodevelopmental delays.
Findings from their study were published in Genetics in Medicine.
“We are so excited about the novel gene discovery,” said the lead author Suma Shankar, professor in the Departments of Pediatrics and Ophthalmology and faculty at the UC Davis MIND Institute. Shankar is the director of Precision Genomics, Albert Rowe Endowed Chair in Genetics, and chief of Division of Genomic Medicine.
Neurons specialized in encoding sound emerge before birth
Distinct neuron types in the auditory organ are necessary for encoding different features of sound and relaying them to the brain. Researchers at Karolinska Institutet provide evidence of an early, neuronal activity-independent, emergence of the different subtypes of auditory neurons, prior to birth in mice. The findings have recently been published in Nature Communications.
Previous studies have provided ambiguous results on whether the different subtypes of auditory neurons emerge during prenatal or postnatal development, with in the latter case, a possible role of neuronal activity in generating their diversity. In this new study, researchers demonstrate that the fate of auditory neuron subtypes is under genetic control in the prenatal period, and reveal the complex molecular networks controlling their genesis.

Proof that Mendel discovered the laws of inheritance decades ahead of his time
Gregor Mendel, the Moravian monk, was indeed “decades ahead of his time and truly deserves the title of ‘founder of genetics.’” So concludes an international team of scientists as the 200th birthday of Mendel approaches on 20 July.
The team, from KeyGene in the Netherlands and the John Innes Centre in the UK, draw on newly-discovered historical information to conclude that, when his proposals are viewed in the light of what was known of cells in the mid-19th century, Mendel was decades ahead of his time.
“Uncovering hidden details about Mendel has helped to build a picture of the scientific and intellectual environment in which he worked. At the outset Mendel knew nothing about genetics and had to deduce it all for himself. How he went about this is highly instructive,” said Dr. Noel Ellis from the John Innes Centre, one of the contributors to the study.
Imagining the Future: The Transformation of Humanity | Peter Diamandis | TEDxLA
In this landmark talk, Peter Diamandis shares how we are rapidly heading towards a human-scale transformation, the next evolutionary step into what he calls a “Meta-Intelligence,” a future in which we are all highly connected — brain to brain via the cloud — sharing thoughts, knowledge and actions.
He highlights the 4 driving forces as well as the 4 steps that is transforming humanity.
In 2014 Fortune Magazine named Peter Diamandis as one of the World’s 50 Greatest Leaders.
Diamandis He is the Founder & Executive Chairman of the XPRIZE Foundation which leads the world in designing and operating large-scale incentive competitions. He is also the Co-Founder & Exec Chairman of Singularity University, a graduate-level Silicon Valley institution that counsels the world’s leaders on exponentially growing technologies.
As an entrepreneur, Diamandis has started 17 companies. He is the Co-Founder and Vice-Chairman of Human Longevity Inc. (HLI), a genomics and cell therapy-based company focused on extending the healthy human lifespan, and Co-Founder and Co-Chairman of Planetary Resources, a company designing spacecraft to enable the detection and prospecting of asteroid for fuels and precious materials.
Peter Diamandis earned degrees in Molecular Genetics and Aerospace Engineering from the MIT, and holds an M.D. from Harvard Medical School.
This talk was given at a TEDx event using the TED conference format but independently organized by a local community.
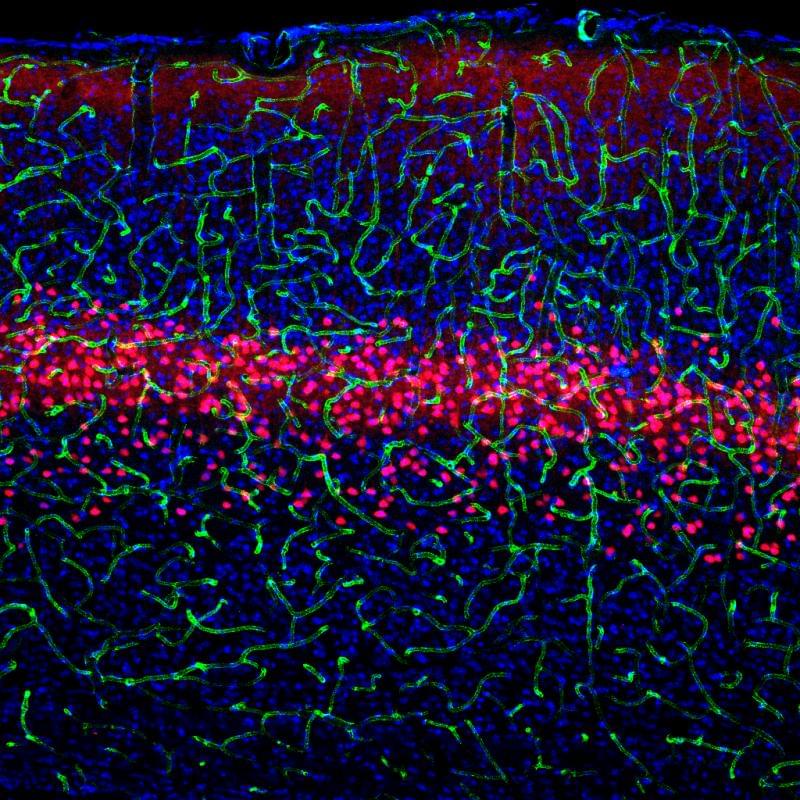
Scientists discover the function and connections of three cell types in the brain
How the brain functions is still a black box: scientists aren’t even sure how many kinds of nerve cells exist in the brain. To know how the brain works, they need to know not only what types of nerve cells exist, but also how they work together. Researchers at the Salk Institute have gotten one step closer to unlocking this black box.
Using cutting-edge visualization and genetic techniques, the team uncovered a new subtype of nerve cell, or neuron, in the visual cortex. The group also detailed how the new cell and two similar neurons process images and connect to other parts of the brain. Learning how the brain analyzes visual information at such a detailed level may one day help doctors understand elements of disorders like schizophrenia and autism.
“Understanding these cell types contributes another piece to the puzzle uncovering neural circuits in the brain, circuits that will ultimately have implications for neurological disorders,” says Edward Callaway, Salk professor and senior author of the paper published December 6 in the journal Neuron.