Circa 2019 immortality in the human brain 🧠
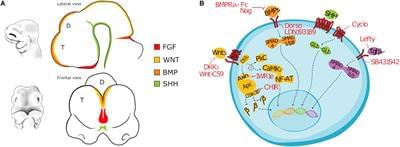
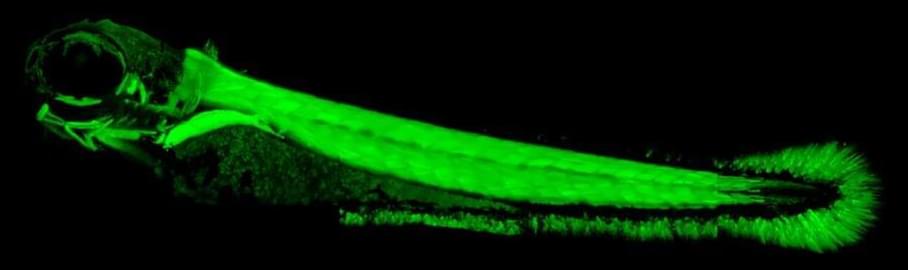
Brain injuries causing chronic sensory or motor deficit, such as stroke, are among the leading causes of disability worldwide, according to the World Health Organization; furthermore, they carry heavy social and economic burdens due to decreased quality of life and need of assistance. Given the limited effectiveness of rehabilitation, novel therapeutic strategies are required to enhance functional recovery. Since cell-based approaches have emerged as an intriguing and promising strategy to promote brain repair, many efforts have been made to study the functional integration of neurons derived from pluripotent stem cells (PSCs), or fetal neurons, after grafting into the damaged host tissue. PSCs hold great promises for their clinical applications, such as cellular replacement of damaged neural tissues with autologous neurons. They also offer the possibility to create in vitro models to assess the efficacy of drugs and therapies. Notwithstanding these potential applications, PSC-derived transplanted neurons have to match the precise sub-type, positional and functional identity of the lesioned neural tissue. Thus, the requirement of highly specific and efficient differentiation protocols of PSCs in neurons with appropriate neural identity constitutes the main challenge limiting the clinical use of stem cells in the near future. In this Review, we discuss the recent advances in the derivation of telencephalic (cortical and hippocampal) neurons from PSCs, assessing specificity and efficiency of the differentiation protocols, with particular emphasis on the genetic and molecular characterization of PSC-derived neurons. Second, we address the remaining challenges for cellular replacement therapies in cortical brain injuries, focusing on electrophysiological properties, functional integration and therapeutic effects of the transplanted neurons.
Brain injuries represent a large variety of disabling pathologies. They may originate from different causes and affect distinct brain locations, leading to an enormous multiplicity of various symptoms ranging from cognitive deficits to sensorimotor disabilities. They can also result in secondary disturbances, such as epileptic foci, which occur within the lesioned and perilesional tissues (Herman, 2002). Indeed, frequently a secondary functional damage can take place in a region distant from the first insult (e.g., the hippocampus after traumatic brain injury), providing an explanation for cognitive and memory deficits arising after a brain lesion (Girgis et al., 2016). Brain injuries can have traumatic or non-traumatic etiologies, including focal brain lesions, anoxia, tumors, aneurysms, vascular malformations, encephalitis, meningitis and stroke (Teasell et al., 2007). In particular, stroke covers a vast majority of acquired brain lesions.