A study of a genetic mouse model of schizophrenia supports two long-debated hypotheses, and unveils additional new clues about the biological roots of the disorder.


Mathematicians delight in the beauty of math that so many of us don’t see. But nature is a wonderful realm in which to observe beauty born out of mathematical relationships.
The natural world provides seemingly endless patterns underpinned by numbers – if we can recognize them.
Luckily for us, a motley team of researchers has just uncovered another striking connection between math and nature; between one of the purest forms of mathematics, number theory, and the mechanisms governing the evolution of life on molecular scales, genetics.

The Death of Death is an international bestseller by José Cordeiro and David Wood that claims that “death will be optional by 2045” – or even earlier, if more public and private funds are invested in rejuvenation technologies.
Longevity. Technology: Already available in more than 10 languages, the book provides insight into recent exponential advances in AI, tissue regeneration, stem cell treatment, organ printing, cryopreservation and genetic therapies that, say the authors, offer a realistic chance to solve the problem of the aging of the human body for the first time in human history. In fact, the book’s subtitle is The Scientific Possibility of Physical Immortality and its Moral Defense.
Given that until relatively recently, just mentioning the concept of ‘biological immortality’ was enough to raise eyebrows and with most of the opinion that it should be filed away under ‘science fiction’ or ‘charlatanism’. However, longevity science is advancing at an incredible pace and today there are people who no longer wonder if immortality is possible, but when it will be a reality. We sat down with José Luis Corderio PhD to find out more.
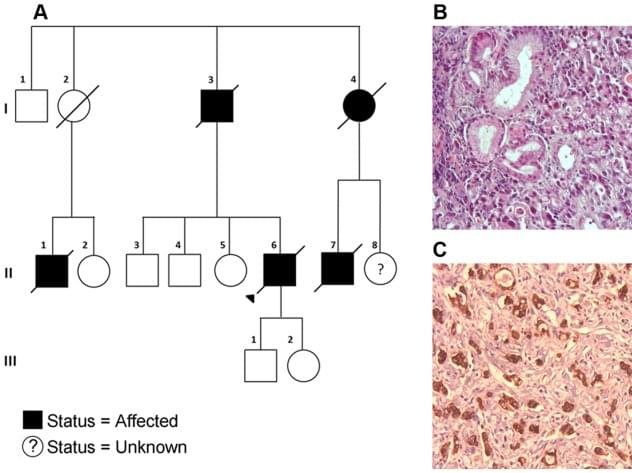
Like biomarker tests, germline testing can help doctors determine the best treatments for patients, but such testing may also help identify people whose family members should be offered testing for potential cancer-causing gene changes.
Guidelines recommend that germline testing be offered to all people with male breast cancer, ovarian cancer, pancreatic cancer, and metastatic prostate cancer. For other cancers with lower likelihood of harmful inherited mutations, recommendations for germline testing vary.
But new findings from a study that is examining the extent of testing for germline mutations among people diagnosed with cancer in California and Georgia between 2013 and 2019 found that germline testing rates are still low. Among the more than 1.3 million people in the study, only about 93,000, or 6.8%, underwent germline genetic testing through March 31, 2021, according to findings published July 3 in JAMA.
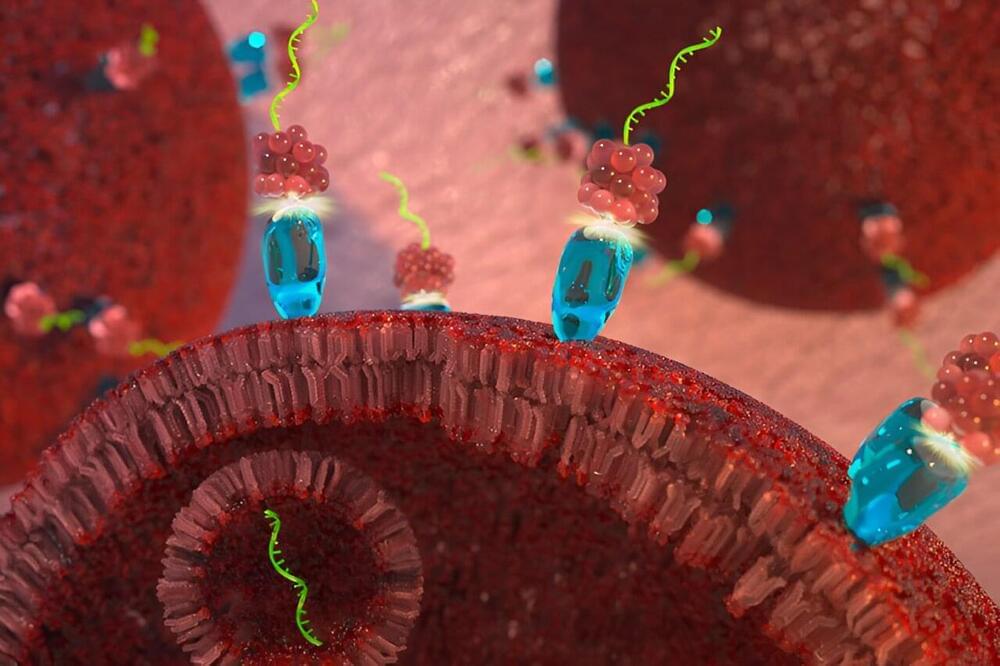
A new cancer therapy developed by Purdue University researchers attacks tumors by tricking cancer cells into absorbing a snippet of RNA that naturally blocks cell division. As reported in Oncogene, tumors treated with the new therapy did not increase in size over the course of a 21-day study, while untreated tumors tripled in size over the same time period. The paper is tiled “A first-in-class fully modified version of miR-34a with outstanding stability, activity, and anti-tumor efficacy.”
Cancer can begin almost anywhere in the human body. It is characterized by cells that divide uncontrollably and that may be able to ignore signals to die or stop dividing, and even evade the immune system. The therapy, tested in mouse models, combines a delivery system that targets cancer cells with a specially modified version of microRNA-34a, a molecule that acts “like the brakes on a car,” slowing or stopping cell division, said Andrea Kasinski, lead author and the William and Patty Miller Associate Professor of biological sciences at Purdue University.
In addition to slowing or reversing tumor growth, the targeted microRNA-34a strongly suppressed the activity of at least three genes—MET, CD44 and AXL—known to drive cancer and resistance to other cancer therapies, for at least 120 hours. The results indicate that the patent-pending therapy, the newest iteration in more than 15 years of work targeting microRNA to destroy cancer, could be effective on its own and in combination with existing drugs when used against cancers that have built drug resistance.

Please see my new FORBES article:
Thanks and please follow me on Linkedin for more tech and cybersecurity insights.
More remarkably, the advent of artificial intelligence (AI) and machine learning-based computers in the next century may alter how we relate to ourselves.
The digital ecosystem’s networked computer components, which are made possible by machine learning and artificial intelligence, will have a significant impact on practically every sector of the economy. These integrated AI and computing capabilities could pave the way for new frontiers in fields as diverse as genetic engineering, augmented reality, robotics, renewable energy, big data, and more.
Three important verticals in this digital transformation are already being impacted by AI: 1) Healthcare, 2) Cybersecurity, and 3) Communications.
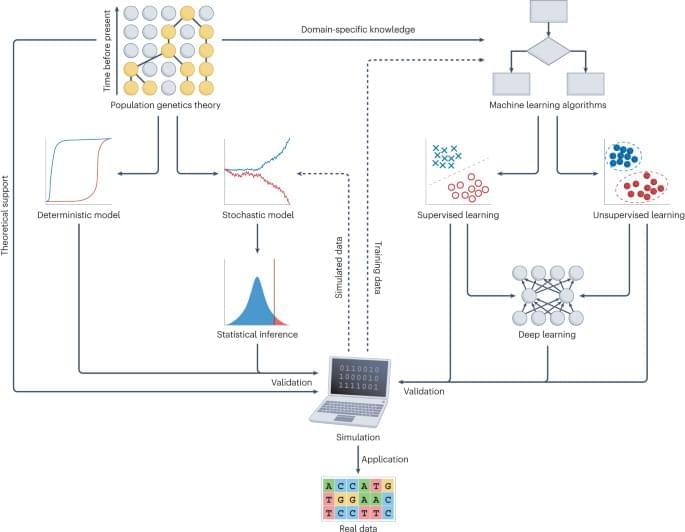
Applying deep learning to large-scale genomic data of species or populations is providing new opportunities to understand the evolutionary forces that drive genetic diversity. This Review introduces common deep learning architectures and provides comprehensive guidelines to implement deep learning models for population genetic inference. The authors also discuss current opportunities and challenges for deep learning in population genetics.


Join us on Patreon! https://www.patreon.com/MichaelLustgartenPhD
Discount Links:
At-Home Metabolomics: https://www.iollo.com?ref=michael-lustgarten.
Use Code: CONQUERAGING At Checkout.
NAD+ Quantification: https://www.jinfiniti.com/intracellular-nad-test/
Use Code: ConquerAging At Checkout.
Epigenetic Testing: https://trudiagnostic.com/?irclickid=U-s3Ii2r7xyIU-LSYLyQdQ6…M0&irgwc=1
Use Code: CONQUERAGING
Oral Microbiome: https://www.bristlehealth.com/?ref=michaellustgarten.
Enter Code: ConquerAging.
At-Home Blood Testing (SiPhox Health): https://getquantify.io/mlustgarten.