Despite their astounding success, custom-made base editors and prime editors will need time to broaden their clinical impact.
You have full access to this article via your institution.
Despite their astounding success, custom-made base editors and prime editors will need time to broaden their clinical impact.
You have full access to this article via your institution.
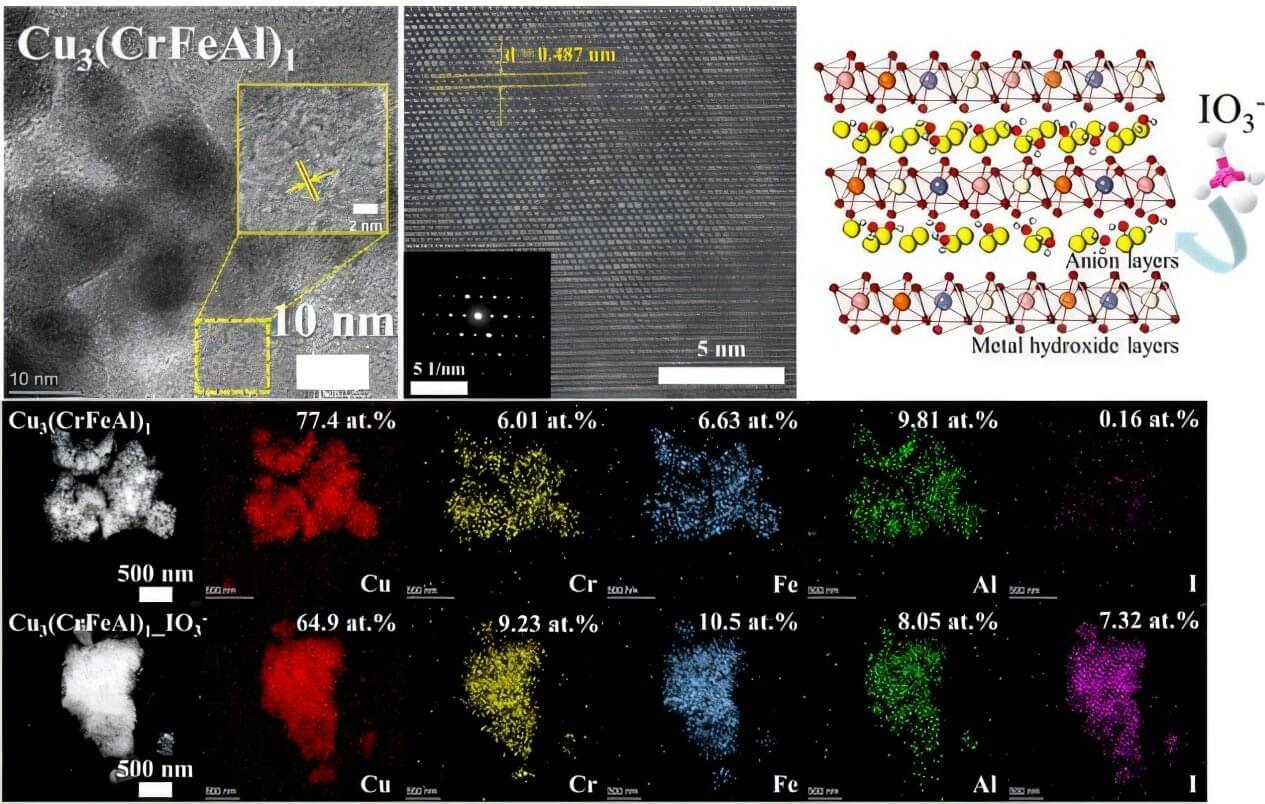
Managing radioactive waste is one of the core challenges in the use of nuclear energy. In particular, radioactive iodine poses serious environmental and health risks due to its long half-life (15.7 million years in the case of I-129), high mobility, and toxicity to living organisms.
A Korean research team has successfully used artificial intelligence to discover a new material that can remove iodine for nuclear environmental remediation. The team plans to push forward with commercialization through various industry–academia collaborations, from iodine-adsorbing powders to contaminated water treatment filters.
Professor Ho Jin Ryu’s research team from the Department of Nuclear and Quantum Engineering, in collaboration with Dr. Juhwan Noh of the Digital Chemistry Research Center at the Korea Research Institute of Chemical Technology, developed a technique using AI to discover new materials that effectively remove radioactive iodine contaminants. Their research is published in the Journal of Hazardous Materials.
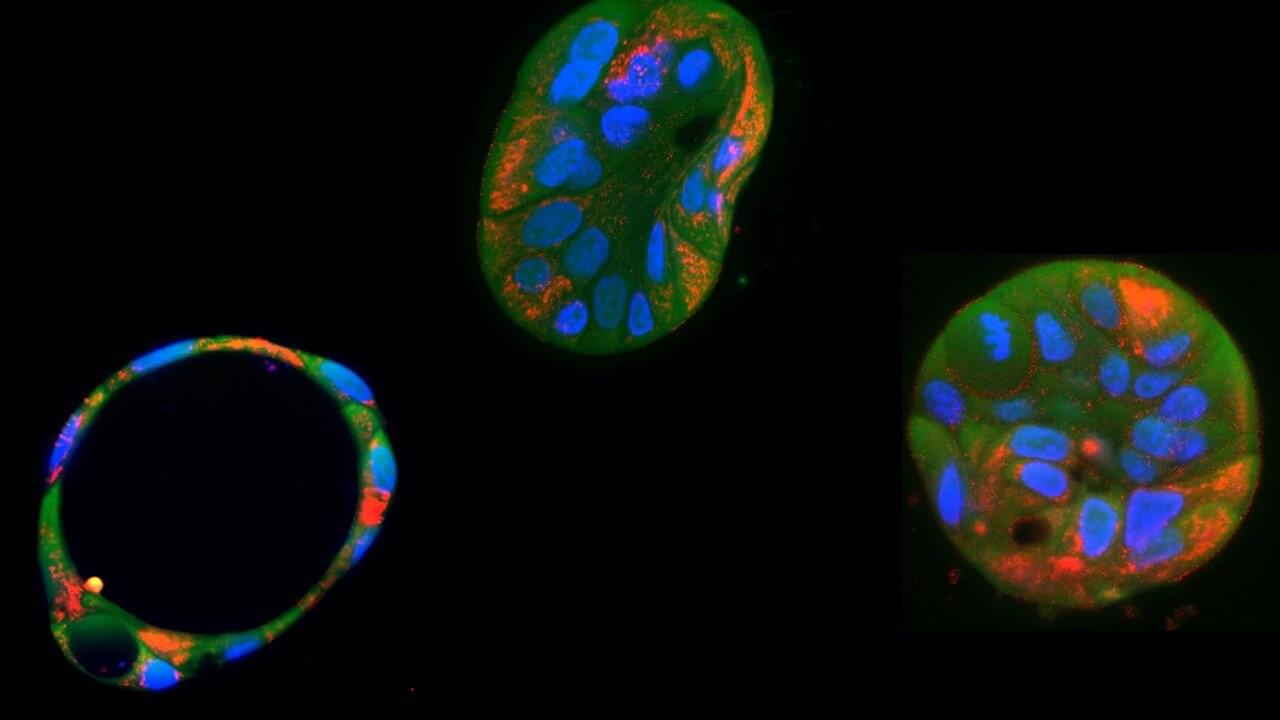
Scientists from QIMR Berghofer’s Cardiac Bioengineering Lab have developed lab-grown, three-dimensional heart tissues known as cardiac organoids that mimic the structure and function of real adult human heart muscle.
To create these tissues, the researchers use special cells called human pluripotent stem cells (which can turn into any cell in the body). However, when these stem cells become heart cells, they usually stay immature and more like the heart tissue found in a developing baby. This immaturity can limit their usefulness to model diseases that present in childhood or as an adult.
In the study, researchers activated two key biological pathways to mimic the effects of exercise in order to mature these cells, making them behave more like genuine adult heart tissue. This breakthrough means scientists can now use these lab-grown heart tissues to test new drugs that could help people with heart conditions. The findings have been published in Nature Cardiovascular Research.
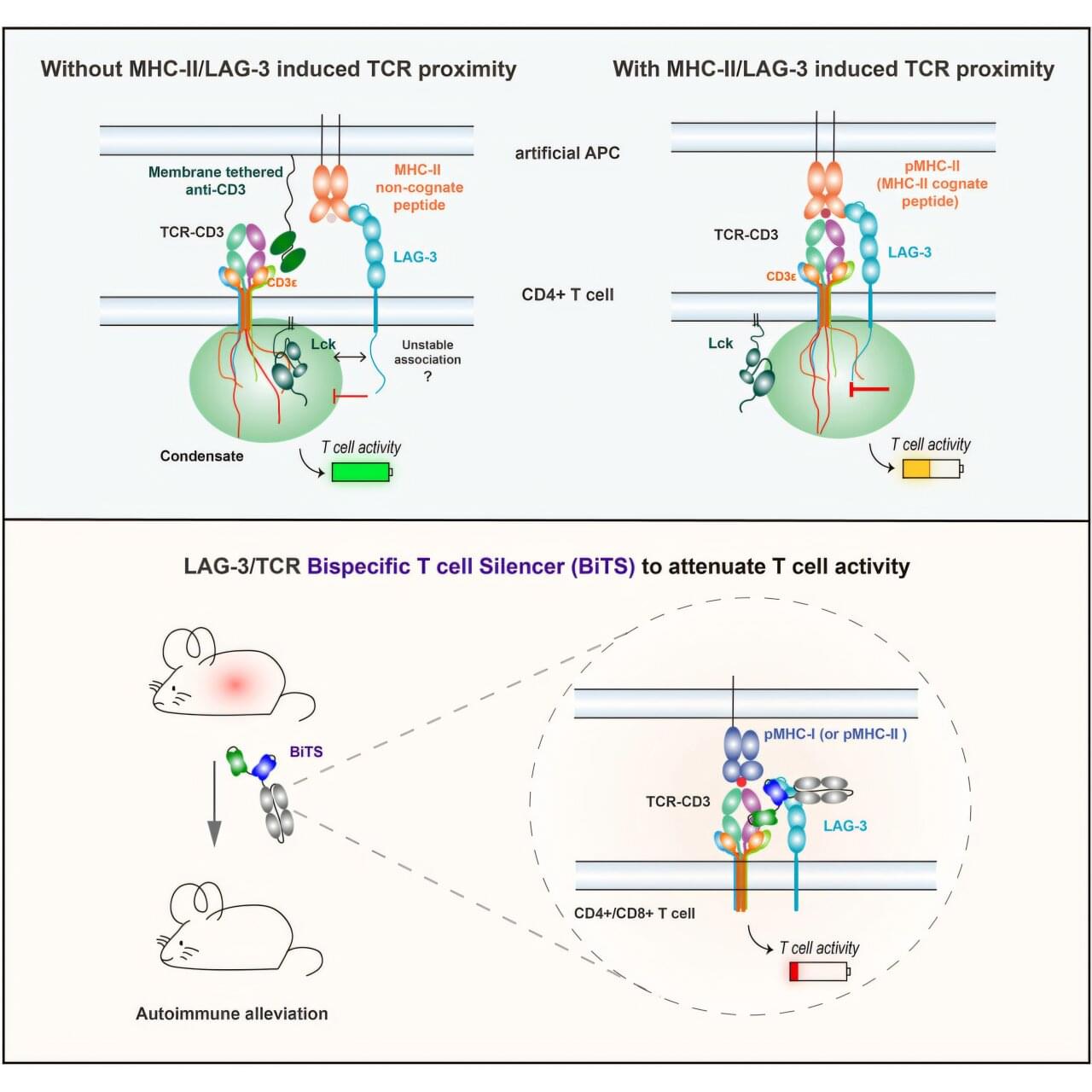
An engineered protein turns off the kind of immune cells most likely to damage tissue as part of type-1 diabetes, hepatitis, multiple sclerosis, shows a new study in mice.
In these autoimmune diseases, T cells mistakenly target the body’s own tissues instead of invading viruses or bacteria as they would during normal immune responses. Treatments focused on T cells have been elusive because blocking their action broadly weakens the immune system and creates risk for infections and cancer.
Published online June 30 in the journal Cell, the study revealed that holding closely together two protein groups (signaling complexes) on T cells, including one found more often on T cells involved in autoimmune disease, shuts down those T cells in a limited way.


In the fight against cancer, immunotherapy—which aims to boost the body’s natural defenses against cancer—is experiencing remarkable growth. Most of these treatments are based on CD8 T lymphocytes, “killer cells” able to eliminate diseased cells. A team from the University of Geneva (UNIGE) has explored an alternative approach involving CD4 T lymphocytes.
Long considered mere auxiliary cells, their therapeutic potential has been considered of secondary importance. But the scientists have discovered that they also have a strong killing capacity, while continuing to support other immune cells. Using cell engineering technologies, the team reprogrammed the cells to target a tumor marker found in many cancers, both in adults and children. These results, published in the journal Science Advances, offer hope for a faster therapeutic strategy that could benefit a greater number of patients.
Traditionally considered as auxiliary cells, CD4 T cells produce molecules to support the action of other immune cells by facilitating their functions, migration or proliferation in the organism. The recent work by Camilla Jandus, Assistant Professor in the Department of Pathology and Immunology, in the Center for Inflammation Research and in the Translational Research Center in Onco-hematology at the UNIGE Faculty of Medicine, shows that they have been vastly underestimated.
Sarcopenia, which is a progressive and extensive decline in muscle mass and strength, is common with aging and estimated to affect up to 50% of people aged 80 and older. It can lead to disability and injuries from falls and is associated with a lower quality of life and an increased mortality. Apart from lifestyle changes, there is no current clinical treatment for sarcopenia.
Space flight with the associated absence of gravity and limited strain on muscles causes muscle weakness, a prominent feature of sarcopenia, within a short period of time, providing a time lapse view on age-related atrophy-associated changes in the muscle. This relatively short window of time in space provides a microgravity model for muscular aging and opens opportunities for studying sarcopenia, which normally takes decades to develop in patients on earth.
To understand the changes of muscle in microgravity, Siobhan Malany, Maddalena Parafati, and their team from the University of Florida, USA, engineered skeletal muscle microtissues from donor biopsies and launched them to the International Space Station (ISS) aboard SpaceX CRS-25. Their findings were published today in Stem Cell Reports. The microtissues were taken from both young, active donors and from aged, sedentary donors and cultured in an automated mini lab, which besides regular feeding and monitoring of cultures also enabled electrical stimulation to simulate exercise. On earth, the contraction strength of microtissues from young, active individuals was almost twice as much as the strength of tissues from older, sedentary individuals. After only two weeks in space, muscle strength trended to decline in the young tissues and was now more comparable to the strength of old tissues. A similar trend was seen for the muscle protein content, which was higher in young microtissues on earth compared to old microtissues but decreased in microgravity to levels measured in old tissues. Further, space flight changed gene expression, particularly in the younger microtissues and disturbed cellular processes related to normal muscle function. Interestingly, electrical stimulation could mitigate these changes in gene expression to some extent.

Gene therapy—once something out of science fiction—is now being used in real hospitals to treat real people. Gene editing has become a conversation of not only treating rare diseases but also about access, fairness, and how much control we should have over our biology.
Genes are sections of DNA that act like instruction manuals telling our cells how to build proteins. Proteins perform vital function like energy use, cellular communications, immunity and cell repair. So when people say “We are what our genes make us,” it’s because these gene-coded proteins guide our growth, health, and behaviour.
Sometimes, typos appear in these instruction manuals. They are called genetic mutations. While many mutations are harmless, some affect the protein made from the mutated gene and disrupt how the cell functions. Some cause serious diseases like cystic fibrosis, muscular atrophy or certain cancers.
Researchers from the Chinese Academy of Sciences and Capital Medical University utilized gene editing to create senescence-resistant human mesenchymal progenitor cells (SRCs). In a 44-week trial on aged macaques, biweekly intravenous SRC injections induced no adverse effects and spurred multi-system rejuvenation in 10 major physiological systems and 61 tissue types. Treated macaques displayed enhanced cognitive function and diminished age-related degeneration. The SRCs work by releasing exosomes that curb cellular senescence and inflammation. This study presents the first primate-level proof of cell therapy’s safety and efficacy in reversing aging, presenting a potential multi-system approach for human anti-aging research.
In a step toward treating mitochondrial diseases, researchers in the Netherlands have successfully edited harmful mutations in mitochondrial DNA using a genetic tool known as a base editor. The results, published in the open-access journal PLOS Biology, offer new hope for people with rare genetic conditions.
Mitochondria have their own small set of DNA. Mutations in this mitochondrial DNA can lead to a wide range of maternally inherited diseases, cancer, and aging-related conditions. While the development of CRISPR technology has given scientists new ways to correct mutations in nuclear DNA, this system cannot effectively cross the mitochondrial membrane and reach mitochondrial DNA.
In the new study, the researchers used a tool called a base editor—specifically, a DdCBE (double-stranded DNA deaminase toxin A-derived cytosine base editor). This tool allows scientists to change a single letter in the DNA code without cutting it, and it works on mitochondrial DNA.