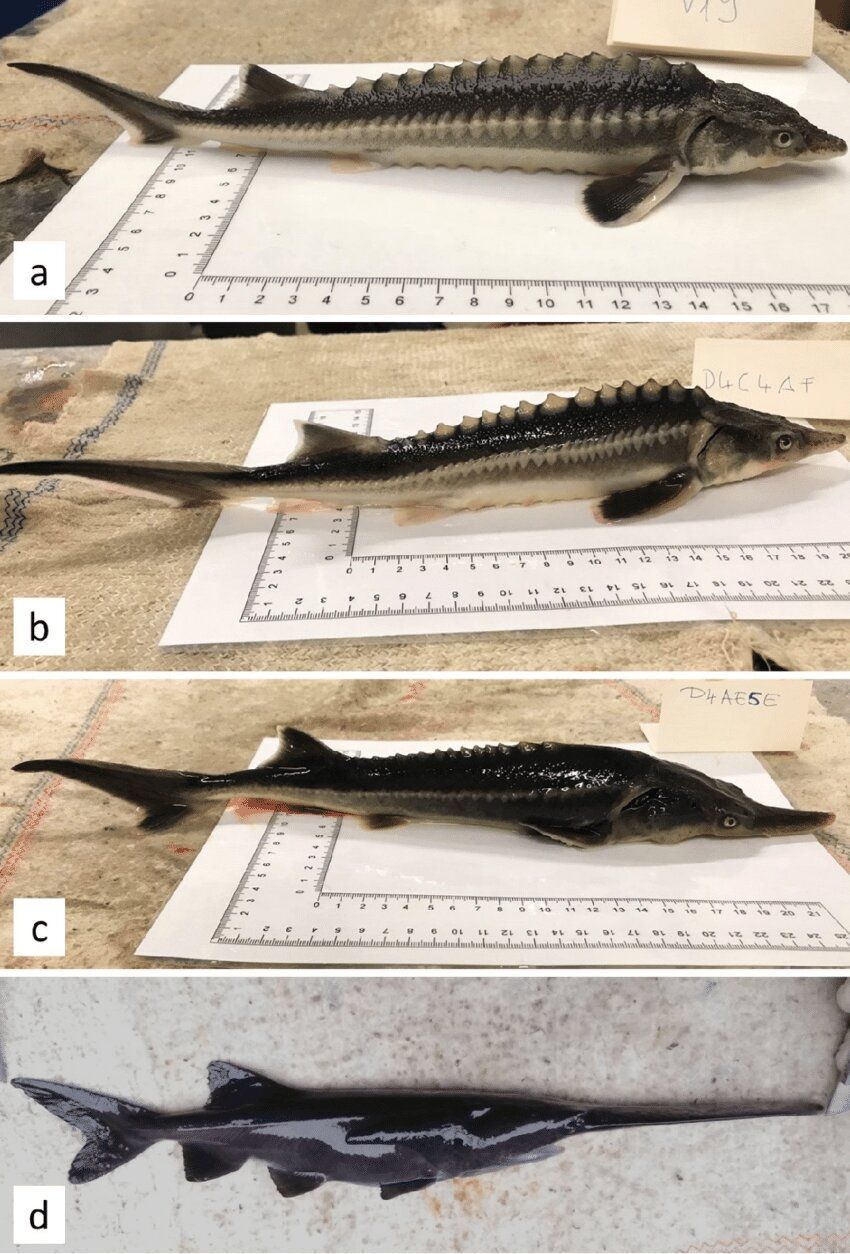
Both shocking and intriguing for the possibilities of gynogenesis reproduction in which sperm is used from one creature to fertilize an egg, but its DNA is ignored.
A team of researchers working at Hungary’s National Agricultural Research and Innovation Centre, Research Institute for Fisheries and Aquaculture, has accidentally bred a new kind of fish—dubbed the sturddlefish by some observers, it is a cross between an American Paddlefish and a Russian Sturgeon. In their paper published in the journal Genes, the group describes accidentally breeding the fish and what they learned by doing so.
In the past, scientists and others have bred animals from different species for various reasons, from research to utility—mules (crossed between donkeys and horses) are considered to have beneficial traits from both animals, and ligers (a cross between lions and tigers) have helped researchers understand their respective genetic backgrounds. In this new effort, the researchers claim that they were not trying to create a new type of fish, they were instead attempting to apply gynogenesis (a type of reproduction in which sperm is used from one creature to fertilize an egg, but its DNA is ignored) using American paddlefish and Russian sturgeon. To their surprise, the eggs produced fish that grew to adults.
In studying the hundreds of offspring produced, which some on the internet have named sturddlefish, the researchers found that they fell into one of three main categories: those that looked mostly like their mothers, those that looked mostly like their fathers and those that inherited features of both parents.