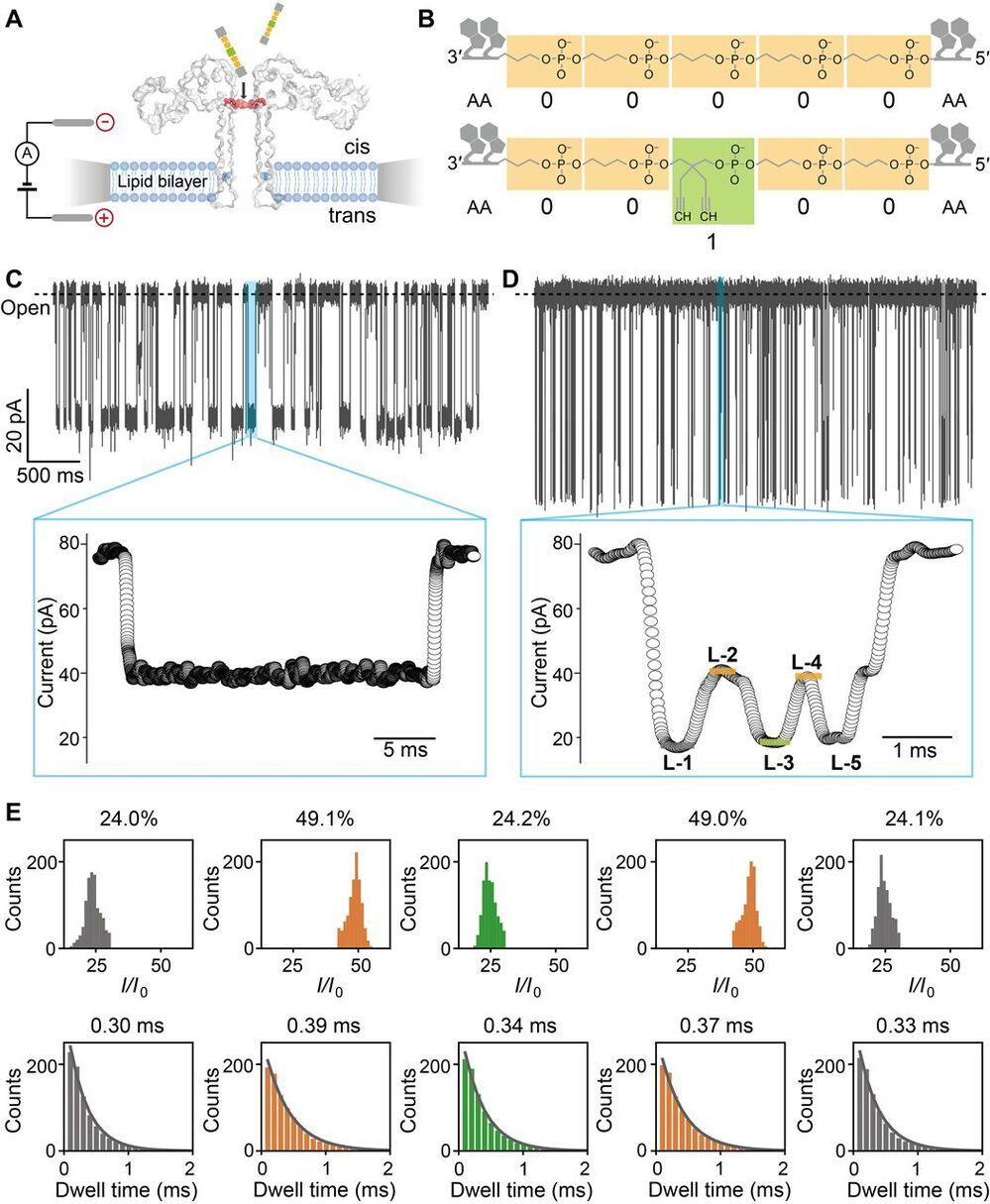
Digital data storage is a growing need for our society and finding alternative solutions than those based on silicon or magnetic tapes is a challenge in the era of “big data.” The recent development of polymers that can store information at the molecular level has opened up new opportunities for ultrahigh density data storage, long-term archival, anticounterfeiting systems, and molecular cryptography. However, synthetic informational polymers are so far only deciphered by tandem mass spectrometry. In comparison, nanopore technology can be faster, cheaper, nondestructive and provide detection at the single-molecule level; moreover, it can be massively parallelized and miniaturized in portable devices. Here, we demonstrate the ability of engineered aerolysin nanopores to accurately read, with single-bit resolution, the digital information encoded in tailored informational polymers alone and in mixed samples, without compromising information density. These findings open promising possibilities to develop writing-reading technologies to process digital data using a biological-inspired platform.
DNA has evolved to store genetic information in living systems; therefore, it was naturally proposed to be similarly used as a support for data storage (1–3), given its high-information density and long-term storage with respect to existing technologies based on silicon and magnetic tapes. Alternatively, synthetic informational polymers have also been described (5–9) as a promising approach allowing digital storage. In these polymers, information is stored in a controlled monomer sequence, a strategy that is also used by nature in genetic material. In both cases, single-molecule data writing is achieved mainly by stepwise chemical synthesis (3, 10, 11), although enzymatic approaches have also been reported (12). While most of the progress in this area has been made with DNA, which was an obvious starting choice, the molecular structure of DNA is set by biological function, and therefore, there is little space for optimization and innovation.