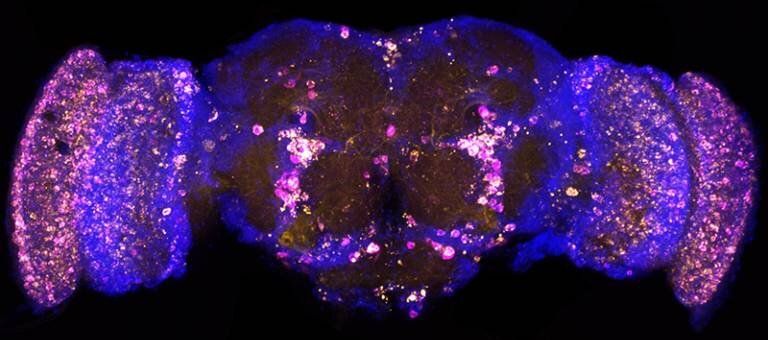
Some genes don’t stay in the same place in the genome. Sometimes called jumping genes or transposons, this genetic material can hop around and rearrange itself | Genetics And Genomics.
Some genetic sequences don’t stay in the same place in the genome. Sometimes called jumping genes or transposons, this genetic material can hop around and rearrange itself to create new sequences. Some transposons even encode for their own enzymes, and these co-called transposases can edit the genome by cutting sequences from one place and pasting them to another.
Reporting in Science, researchers have now suggested that transposable elements (TEs) can fuse with portions of existing genes that code for protein called exons, and get incorporated into genes in a process called exon shuffling to create novel genes that are functional, and express new proteins.