In “The Genetic Lottery: Why DNA Matters for Social Equality,” Kathryn Paige Harden explores how genetics can affect life outcomes.


Track code: TD-3
Abstract:
Solar Sails are at the same stage of engineering development as electric motors were in the 1830’s. Each attribute of solar flux has been examined in isolation, such as photon, proton, plasma, and electrodynamic systems. This talk recommends designing a simple baseline system that converges multiple propulsion methods into optimized systems, as is currently done with electric motors. Many convergences can come from this solution space. Once a baseline design is created, AI genetic algorithms can “flight test” and refine the designs in simulation to adjust proportions and geometry. Once a base design is refined, a second AI evolution pass would design fleet systems that flock like birds to optimize performance. These could fly as a protective shield around Mars crewed fleets, provide space based solar power, deploy rapid reaction probes for interstellar comets, and be used in NEO asteroid mining. In the long term, fleets of solar energy management vehicles can provide orbital Carrigan event protection and Martian solar wind protection for terraforming. This talk is also a case study in how technology revolutions happen, and how to accelerate the creation and democratization of technical solutions.
From the 24th Annual International Mars Society Convention, held as a Virtual Convention worldwide on the Internet from October 14–17, 2021. The four-day International Mars Society Convention, held every year since 1,998 brings together leading scientists, engineers, aerospace industry representatives, government policymakers and journalists to talk about the latest scientific discoveries, technological advances and political-economic developments that could help pave the way for a human mission to the planet Mars.
Conference Papers and some presentations will be available on www.MarsPapers.org.
For more information on the Mars Society, visit our website at www.MarsSociety.org.
#MarsSociety #MarsSocCon2021
Circa 2018
The secrets to immortality may lie in an unexpected place — fruit fly stem cells. Researchers led by Howard Hughes Medical Institute (HHMI) Investigator Yukiko Yamashita have found that some stem cells have a genetic trick to remain young forever across generations. While some areas of the fruit fly genome get shorter as they age, some reproductive cells are able to fix that shortening. Once observed only in yeast, this work, reported in eLife, has revealed more about aging, and how some cells can avoid it.
This work focused on critical genes in ribosomal DNA, rDNA. Ribosomes are cellular organelles that act as protein factories. That rDNA is repeated in several areas of the genome because many ribosomes are needed to make all of the proteins the body needs. Five chromosomes each have spots with hundreds of copies of rDNA. However, that type of redundant sequence can be difficult for cells to replicate accurately every time cell division happens.

An artificial intelligence (AI)-based technology rapidly diagnoses rare disorders in critically ill children with high accuracy, according to a report by scientists from University of Utah Health and Fabric Genomics, collaborators on a study led by Rady Children’s Hospital in San Diego. The benchmark finding, published in Genomic Medicine, foreshadows the next phase of medicine, where technology helps clinicians quickly determine the root cause of disease so they can give patients the right treatment sooner.
“This study is an exciting milestone demonstrating how rapid insights from AI-powered decision support technologies have the potential to significantly improve patient care,” says Mark Yandell, Ph.D., co-corresponding author on the paper. Yandell is a professor of human genetics and Edna Benning Presidential Endowed Chair at U of U Health, and a founding scientific advisor to Fabric.
Worldwide, about seven million infants are born with serious genetic disorders each year. For these children, life usually begins in intensive care. A handful of NICUs in the U.S., including at U of U Health, are now searching for genetic causes of disease by reading, or sequencing, the three billion DNA letters that make up the human genome. While it takes hours to sequence the whole genome, it can take days or weeks of computational and manual analysis to diagnose the illness.
Circa 2019 😀
Because they can process massive amounts of data, computers can perform analytical tasks that are beyond human capability. Google, for instance, is using its computing power to develop AI algorithms that construct two-dimensional CT images of lungs into a three-dimensional lung and look at the entire structure to determine whether cancer is present. Radiologists, in contrast, have to look at these images individually and attempt to reconstruct them in their heads. Another Google algorithm can do something radiologists cannot do at all: determine patients’ risk of cardiovascular disease by looking at a scan of their retinas, picking up on subtle changes related to blood pressure, cholesterol, smoking history and aging. “There’s potential signal there beyond what was known before,” says Google product manager Daniel Tse.
The Black Box Problem
AI programs could end up revealing entirely new links between biological features and patient outcomes. A 2019 paper in JAMA Network Open described a deep-learning algorithm trained on more than 85,000 chest x-rays from people enrolled in two large clinical trials that had tracked them for more than 12 years. The algorithm scored each patient’s risk of dying during this period. The researchers found that 53 percent of the people the AI put into a high-risk category died within 12 years, as opposed to 4 percent in the low-risk category. The algorithm did not have information on who died or on the cause of death. The lead investigator, radiologist Michael Lu of Massachusetts General Hospital, says that the algorithm could be a helpful tool for assessing patient health if combined with a physician’s assessment and other data such as genetics.

Summary: Researchers have linked Fragile X and SHANK3 deletion syndrome, two disorders associated with autism, to specific microscopic walking patterns.
Source: Rutgers.
Rutgers researchers have linked the genetic disorders Fragile X and SHANK3 deletion syndrome – both linked to autism and health problems – to walking patterns by examining the microscopic movements of those wearing motion-sensored sneakers.
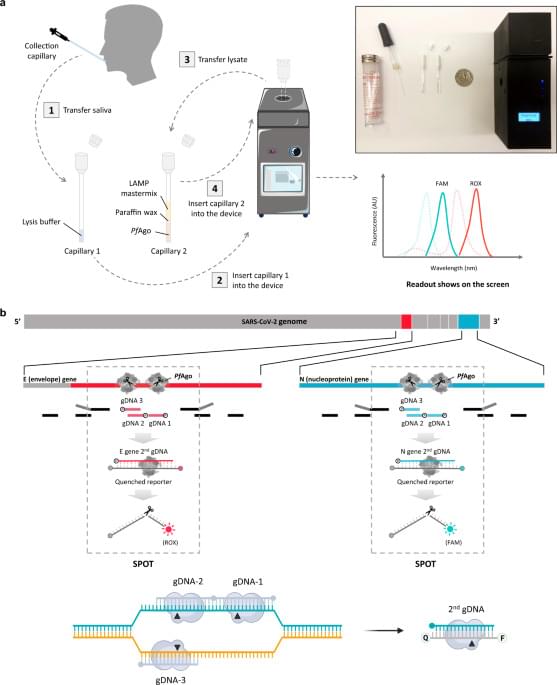
Here we report a rapid Scalable and Portable Testing (SPOT) system consisting of a rapid, highly sensitive, and accurate assay and a battery-powered portable device for COVID-19 diagnosis. This device consists of 3D printed casing and internal structure with precise temperature control and fluorescence detection, whereas this assay combines RT-LAMP with an Argonaute protein from hyperthermophilic archaeon Pyrococcus furiosus (PfAgo) capable of precise recognition and cleavage of a target DNA at 95 °C as directed by small 5′-phosphorylated single strand DNA (ssDNA) as guide DNA (gDNA)10,11. Due to the multi-turnover activity of PfAgo, its secondary cleavage mechanism can be harnessed for specific, sensitive, and multiplex nucleic acid detection12,13. For COVID-19 samples, although nasopharyngeal swab and nasal swab samples were recommended for detection of SARS-CoV-2, saliva samples are a more attractive alternative due to the ease, safety, and non-invasive nature of its collection14,15, and its relatively high viral load during the first week of infection16. These benefits enable a saliva sample to be an ideal specimen for reliable and rapid self-detection without professional supervision17,18,19. While current CRISPR-based detection systems normally require 50 min for testing, PfAgo can dramatically speed up the detection process by requiring only 3–5 min for cleavage of amplified products, thereby shortening the total turnaround time for testing to less than 30 min. Moreover, successful PfAgo detection requires at least two sequence-specific cleavages, endowing the SPOT system with high specificity and the ability for multiplexing. Finally, to validate the SPOT system, the sensitivity and accuracy of the SPOT system were determined by using 104 clinical saliva samples.
- please note: The SPOT system also may be useful for detecting genetic markers of certain types of cancer in saliva.
There is a clear need for rapid, accurate and scalable Covid-19 diagnostics. Here the authors use PfAgo to detect viral sequences amplified by RT-LAMP in a handheld battery-powered device.



A team of researchers at UT Southwestern Medical Center’s Touchstone Diabetes Center have successfully used CRISPR gene editing to turn fat cells normally used for storage into energy-burning cells.
“It’s like flipping a switch. We removed the ‘brake’ on the energy burning pathway in fat cells by engineering a mutation that disrupts the interaction between a single pair of proteins,” said study leader Rana Gupta, Ph.D., Associate Professor of Internal Medicine. “Our research demonstrates that releasing this brake in fat cells can potentially help make existing diabetes medications much more effective.”
The research at UT Southwestern, ranked as one of the nation’s top 25 hospitals for diabetes and endocrinology care, is published in Genes and Development and supported by the National Institutes of Health.