Realistic and complex models of brain cells, developed at Cedars-Sinai with support from our scientists and our #openscience data, could help answer questions a… See more.
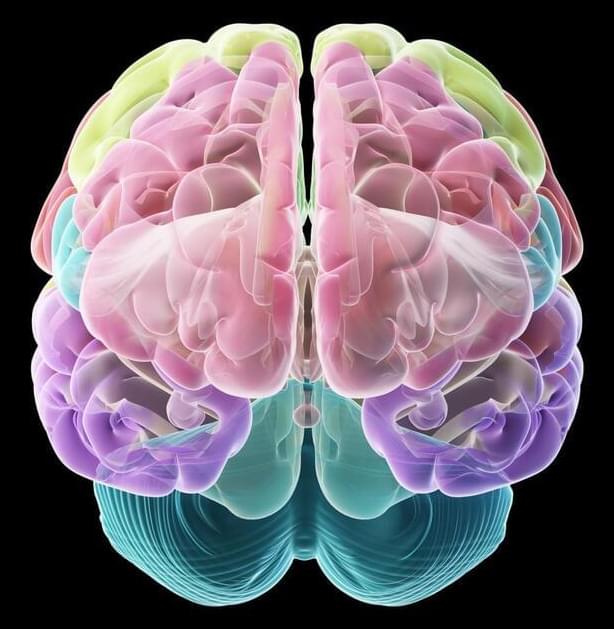
Cedars-Sinai investigators have created bio-realistic and complex computer models of individual brain cells—in unparalleled quantity.
Their research, published today in the peer-reviewed journal Cell Reports, details how these models could one day answer questions about neurological disorders—and even human intellect—that aren’t possible to explore through biological experiments.
“These models capture the shape, timing and speed of the electrical signals that neurons fire in order to communicate with each other, which is considered the basis of brain function,” said Costas Anastassiou, PhD, a research scientist in the Department of Neurosurgery at Cedars-Sinai, and senior author of the study. “This lets us replicate brain activity at the single-cell level.”
The models are the first to combine data sets from different types of laboratory experiments to present a complete picture of the electrical, genetic and biological activity of single neurons. The models can be used to test theories that would require dozens of experiments to examine in the lab, Anastassiou said.