Groundbreaking maps reveal the complex gene regulation in brains with and without mental disorders, enhancing the understanding of mental illnesses and potential treatments.
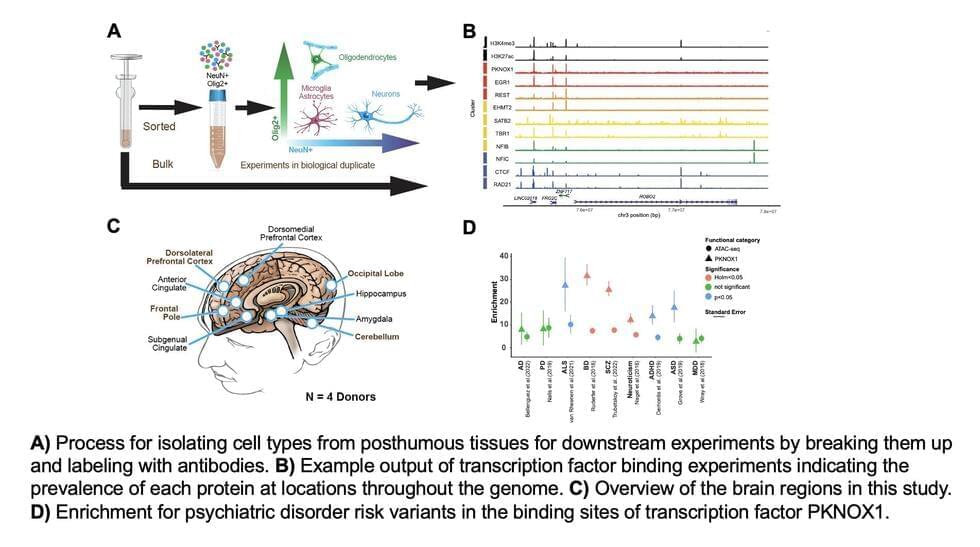
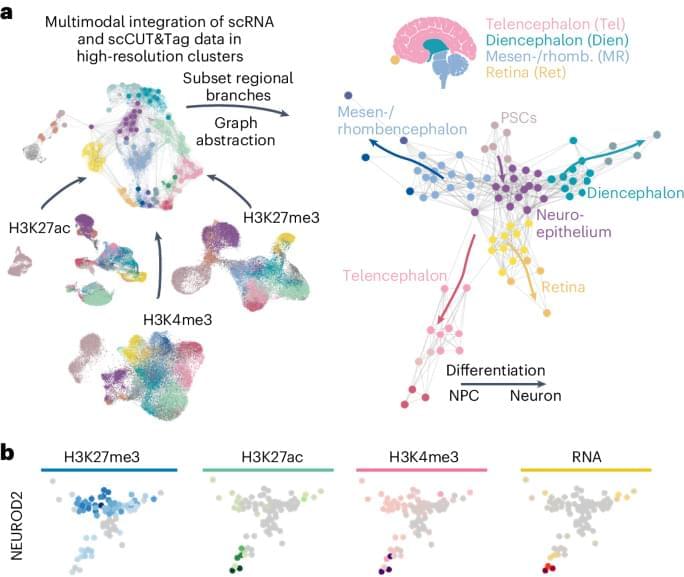
A consortium of researchers has produced the largest and most advanced multidimensional maps of gene regulation networks in the brains of people with and without mental disorders. These maps detail the many regulatory elements that coordinate the brain’s biological pathways and cellular functions. The research, supported by the National Institutes of Health (NIH), used postmortem brain tissue from over 2,500 donors to map gene regulation networks across different stages of brain development and multiple brain-related disorders.
“These groundbreaking findings advance our understanding of where, how, and when genetic risk contributes to mental disorders such as schizophrenia, post-traumatic stress disorder, and depression,” said Joshua A. Gordon, M.D., Ph.D., director of NIH’s National Institute of Mental Health (NIMH). “Moreover, the critical resources, shared freely, will help researchers pinpoint genetic variants that are likely to play a causal role in mental illnesses and identify potential molecular targets for new therapeutics.”