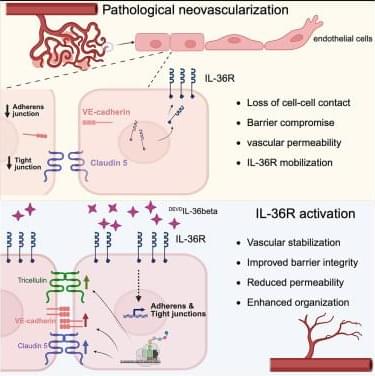
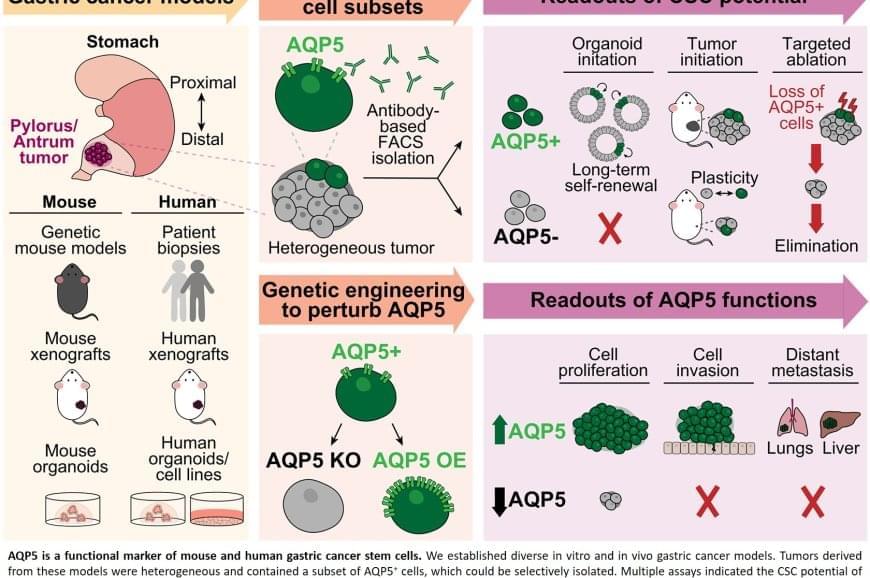
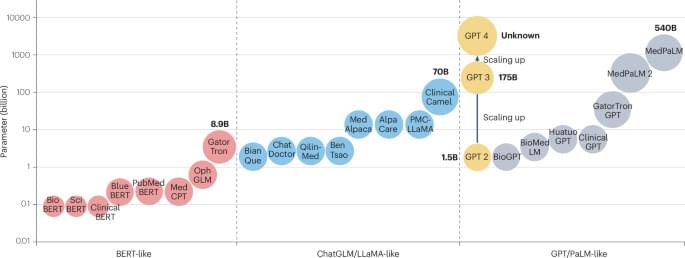
The device is also used in selected neurological cases where accurate signal detection and responsive stimulation are critical to managing symptoms over time. Its application forms part of a broader treatment pathway rather than a standalone intervention.
Implantation is performed using minimally invasive techniques and typically takes three to five hours. The approach avoids large surgical incisions and supports shorter recovery periods, allowing patients to resume daily activities more quickly.
Rather than marking a single milestone, the continued use of this technology reflects KFSHRC’s integration of artificial intelligence into routine neurological care, where adaptability and long-term management are central to patient outcomes.
RIYADH, SAUDI ARABIA, December 28, 2025 /EINPresswire.com/ — At King Faisal Specialist Hospital & Research Centre (KFSHRC) in Riyadh, artificial intelligence enabled brain implants are used as part of advanced care for patients with neurological conditions, including Parkinson’s disease and selected movement disorders.
The implant functions by continuously analyzing brain signals and responding to abnormal activity through targeted electrical stimulation. This adaptive approach allows treatment to adjust in real time based on the patient’s neural patterns, reducing reliance on fixed stimulation settings and limiting the need for frequent manual recalibration.
In clinical practice, the technology has supported improved symptom control for patients whose conditions require precise neuromodulation. As treatment progresses, some patients have been able to reduce their dependence on medication under clinical supervision, while maintaining daily function and stability.