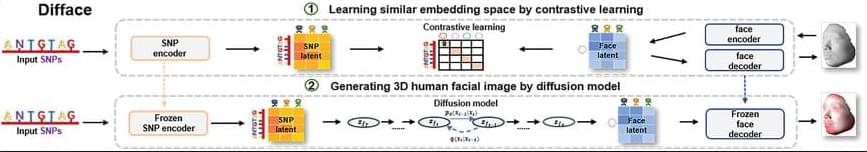
Facial morphology is a distinctive biometric marker, offering invaluable insights into personal identity, especially in forensic science. In the context of high-throughput sequencing, the reconstruction of 3D human facial images from DNA is becoming a revolutionary approach for identifying individuals based on unknown biological specimens. Inspired by artificial intelligence techniques in text-to-image synthesis, it proposes Difface, a multi-modality model designed to reconstruct 3D facial images only from DNA. Specifically, Difface first utilizes a transformer and a spiral convolution network to map high-dimensional Single Nucleotide Polymorphisms and 3D facial images to the same low-dimensional features, respectively, while establishing the association between both modalities in the latent features in a contrastive manner; and then incorporates a diffusion model to reconstruct facial structures from the characteristics of SNPs. Applying Difface to the Han Chinese database with 9,674 paired SNP phenotypes and 3D facial images demonstrates excellent performance in DNA-to-3D image alignment and reconstruction and characterizes the individual genomics. Also, including phenotype information in Difface further improves the quality of 3D reconstruction, i.e. Difface can generate 3D facial images of individuals solely from their DNA data, projecting their appearance at various future ages. This work represents pioneer research in de novo generating human facial images from individual genomics information.
(Repost)
This study has introduced Difface, a de novo multi-modality model to reconstruct 3D facial images from DNA with remarkable precision, by a generative diffusion process and a contrastive learning scheme. Through comprehensive analysis and SNP-FACE matching tasks, Difface demonstrated superior performance in generating accurate facial reconstructions from genetic data. In particularly, Difface could generate/predict 3D facial images of individuals solely from their DNA data at various future ages. Notably, the model’s integration of transformer networks with spiral convolution and diffusion networks has set a new benchmark in the fidelity of generated images to their real images, as evidenced by its outstanding accuracy in critical facial landmarks and diverse facial feature reproduction.
Difface’s novel approach, combining advanced neural network architectures, significantly outperforms existing models in genetic-to-phenotypic facial reconstruction. This superiority is attributed to its unique contrastive learning method of aligning high-dimensional SNP data with 3D facial point clouds in a unified low-dimensional feature space, a process further enhanced by adopting diffusion networks for phenotypic characteristic generation. Such advancements contribute to the model’s exceptional precision and ability to capture the subtle genetic variations influencing facial morphology, a feat less pronounced in previous methodologies.
Despite Difface’s demonstrated strengths, there remain directions for improvement. Addressing these limitations will require a focused effort to increase the model’s robustness and adaptability to diverse datasets. Future research should aim to incorporating variables like age and BMI would allow Difface to simulate age-related changes, enabling the generation of facial images at different life stages an application that holds significant potential in both forensic science and medical diagnostics. Similarly, BMI could help the model account for variations in body composition, improving its ability to generate accurate facial reconstructions across a range of body types.