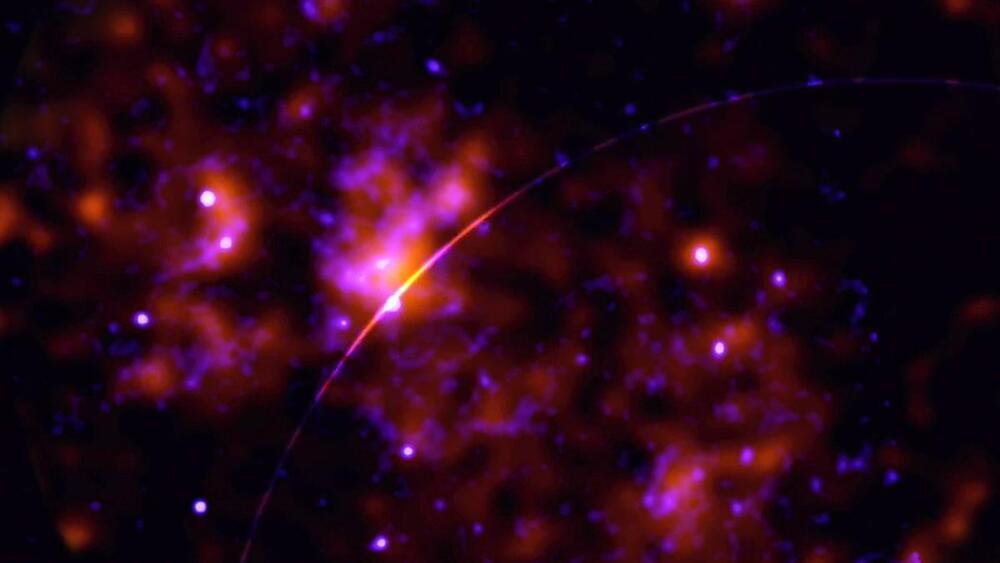
After a search of neutron stars finds preliminary evidence for hypothetical dark matter particles called axions, astrophysicists are devising new ways to spot them.



How much water a fish consumes really depends on how much salt is in its surrounding habitat. While fish do drink some water — salty or fresh, depending on their surroundings — through their mouths, they mostly absorb it through their skin and gills via osmosis.
“You’ve got to think of a fish as sort of a leaky boat in the water,” Tim Grabowski, a marine biologist at the University of Hawaii, told Live Science. “You constantly have a movement of either water or the salts that are in the water between the fish’s body and the external environment.”

Scandinavian studios Henning Larsen and White Arkitekter are designing Stockholm Wood City, which will become the world’s largest mass-timber development and have the “serenity of a forest”.
Set to be built in the Stockholm neighbourhood of Sickla, the project was dubbed the “world’s largest wooden city” by developer Atrium Ljungberg as it will use more timber that any other project in development.
Stockholm Wood City, which will have 7,000 office spaces and 2,000 homes and cover 250,000 square metres, is being designed by Danish studio Henning Larsen and Swedish firm White Arkitekter.
O.o!!!! Year 2022
A new photonic quantum computer takes just 36 microseconds to perform a task that would take a conventional supercomputer more than 9,000 years to complete. The new device, named Borealis, is the first quantum computer from a startup to display such “quantum advantage” over regular computers. Borealis is also the first machine capable of quantum advantage to be made available to the public over the cloud.
Quantum computers can theoretically achieve a quantum advantage that enables them to find the answers to problems no classical computers could ever solve. The more components known as qubits that a quantum computer has, the greater its computational power can grow, in an exponential fashion.
Many companies, including giants such as Google, IBM, and Amazon as well as startups such as IonQ, rely on qubits based on superconducting circuits or trapped ions. One drawback with these approaches is that they both demand temperatures colder than those found in deep space, because heat can disrupt the qubits. The expensive, bulky cryogenic systems required to hold qubits at such frigid temperatures can also make it a major challenge to scale these platforms up to high numbers of qubits—or to smaller and more portable form factors.


Apple today introduced the first version of the visionOS software, debuting the visionOS 1.0 Developer Beta. The introduction of the beta comes as Apple has announced the launch of the visionOS software development kit (SDK) that will allow third-party developers to build apps for the Vision Pro headset.
The SDK can be accessed through Xcode 15 beta 2, and while developers do not have access to the Vision Pro headset itself as of yet, Apple will begin allowing testing starting next month.
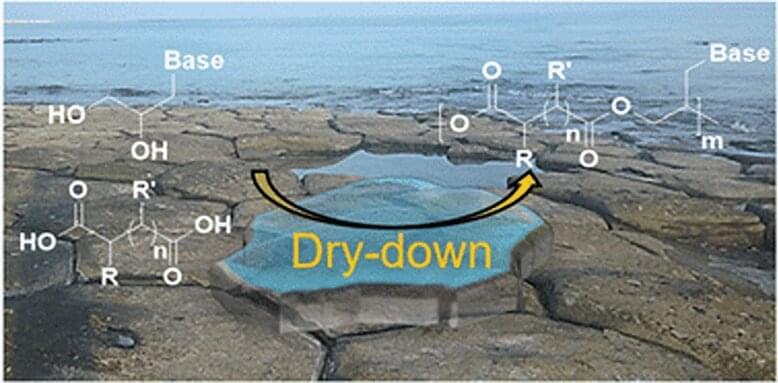
The chemical origin of life on Earth is a puzzle that scientists have been trying to piece together for decades. Many hypotheses have been proposed to explain how life came to be and what chemical and environmental factors on early Earth could have led to it. A step required in a number of these hypotheses involves the abiotic synthesis of genetic polymers—materials made up of a sequence of repeating chemical units with the ability to store and pass down information through base-pairing interactions.
One such hypothesis is the RNA (ribonucleic acid) world hypothesis, which draws from this concept and suggests that RNA could have been the original biopolymer of life both for genetic information storage and transmission, and for catalysis. However, in the absence of chemical activation of RNA monomers, studies have found that RNA polymerization would have been inefficient under primitive dry-down conditions without specialized circumstances such as lipid or salt-assisted synthesis or mineral templating.
While this does not necessarily make the RNA world hypothesis less plausible, primitive chemical systems were quite diverse and could not have possibly been as clean to just contain RNA and lipids, suggesting that other forms of primitive nucleic acid polymerization may have also taken place.

The discovery of the quantum Hall effects in the 1980s revealed the existence of novel states of matter called “Laughlin states,” in honor of the American Nobel prize winner who successfully characterized them theoretically. These exotic states specifically emerge in 2D materials, at very low temperature and in the presence of an extremely strong magnetic field.
In a Laughlin state, electrons form a peculiar liquid, where each electron dances around its congeners while avoiding them as much as possible. Exciting such a quantum liquid generates collective states that physicists associate with fictitious particles, whose properties drastically differ from electrons: these “anyons” carry a fractional charge (a fraction of the elementary charge) and they surprisingly defy the standard classification of particles in terms of bosons or fermions.
For many years, physicists have explored the possibility of realizing Laughlin states in other types of systems than those offered by solid-state materials, in view of further analyzing their peculiar properties. However, the required ingredients (the 2D nature of the system, the intense magnetic field, the strong correlations among the particles) has proved extremely challenging.
