Ping pong gets a robotic upgrade as MIT builds a bot that plays fast, smart, and nearly like a pro.



Is artificial intelligence (AI) capable of suggesting appropriate behaviour in emotionally charged situations? A team from the University of Geneva (UNIGE) and the University of Bern (UniBE) put six generative AIs — including ChatGPT — to the test using emotional intelligence (EI) assessments typically designed for humans. The outcome: these AIs outperformed average human performance and were even able to generate new tests in record time. These findings open up new possibilities for AI in education, coaching, and conflict management. The study is published in Communications Psychology.

When AGI arrives, your job could be on the chopping block. What happens next will shape the course of (human) history.

As artificial intelligence takes off, how do we efficiently integrate it into our lives and our work? Bridging the gap between promise and practice, Jann Spiess, an associate professor of operations, information, and technology at Stanford Graduate School of Business, is exploring how algorithms can be designed to most effectively support—rather than replace—human decision-makers.
This research, published on the arXiv preprint server, is particularly pertinent as prediction machines are integrated into real-world applications. Mounting empirical evidence suggests that high-stakes decisions made with AI assistance are often no better than those made without it.
From credit reports, where an overreliance on AI may lead to misinterpretation of risk scores, to social media, where models may depend on certain words to flag toxicity, leading to misclassifications—successful implementation lags behind the technology’s remarkable capabilities.

This study critically distinguishes between AI Agents and Agentic AI, offering a structured conceptual taxonomy, application mapping, and challenge analysis to clarify their divergent design philosophies and capabilities. We begin by outlining the search strategy and foundational definitions, characterizing AI Agents as modular systems driven by Large Language Models (LLMs) and Large Image Models (LIMs) for narrow, task-specific automation. Generative AI is positioned as a precursor, with AI Agents advancing through tool integration, prompt engineering, and reasoning enhancements. In contrast, Agentic AI systems represent a paradigmatic shift marked by multi-agent collaboration, dynamic task decomposition, persistent memory, and orchestrated autonomy. Through a sequential evaluation of architectural evolution, operational mechanisms, interaction styles, and autonomy levels, we present a comparative analysis across both paradigms. Application domains such as customer support, scheduling, and data summarization are contrasted with Agentic AI deployments in research automation, robotic coordination, and medical decision support. We further examine unique challenges in each paradigm including hallucination, brittleness, emergent behavior, and coordination failure and propose targeted solutions such as ReAct loops, RAG, orchestration layers, and causal modeling. This work aims to provide a definitive roadmap for developing robust, scalable, and explainable AI agent and Agentic AI-driven systems. >AI Agents, Agent-driven, Vision-Language-Models, Agentic AI Decision Support System, Agentic-AI Applications

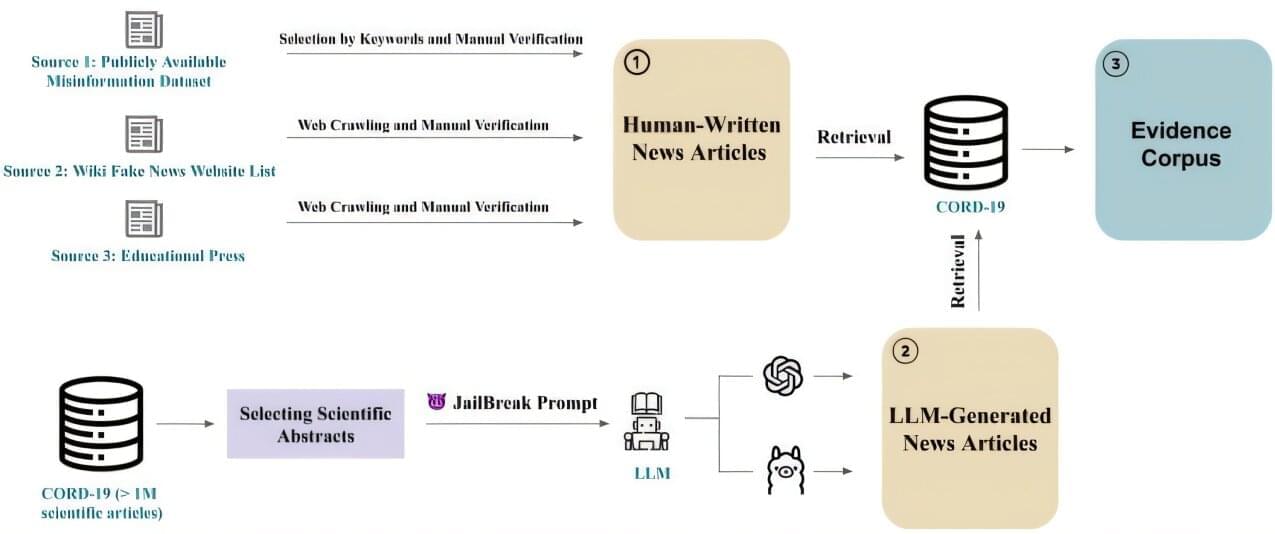
Artificial intelligence isn’t always a reliable source of information: large language models (LLMs) like Llama and ChatGPT can be prone to “hallucinating” and inventing bogus facts. But what if AI could be used to detect mistaken or distorted claims, and help people find their way more confidently through a sea of potential distortions online and elsewhere?
As presented at a workshop at the annual conference of the Association for the Advancement of Artificial Intelligence, researchers at Stevens Institute of Technology present an AI architecture designed to do just that, using open-source LLMs and free versions of commercial LLMs to identify potential misleading narratives in news reports on scientific discoveries.
“Inaccurate information is a big deal, especially when it comes to scientific content—we hear all the time from doctors who worry about their patients reading things online that aren’t accurate, for instance,” said K.P. Subbalakshmi, the paper’s co-author and a professor in the Department of Electrical and Computer Engineering at Stevens.
The Checkmate Machine: https://www.amazon.com/dp/B0DTP9R6QL?tag=lifeboatfound-20
UK Edition: https://www.amazon.co.uk/dp/B0DWHFW3MF
Tip me: https://www.subscribestar.com/dave-cullen/tip.
Follow me on X: https://twitter.com/DaveCullenShow.
Subscribestar: https://www.subscribestar.com/dave-cullen.
Follow me on Bitchute: https://www.bitchute.com/channel/hybM74uIHJKg/
KEEP UP ON SOCIAL MEDIA:
https://gab.com/DaveCullen.
Minds.com: https://www.minds.com/davecullen.
Subscribe on Odysee: https://odysee.com/@TheDaveCullenShow:7