Are you running a small business? Learn how to use AI for small businesses effectively and what tools to use to get the most out of the AI.


This post is also available in:  עברית (Hebrew)
עברית (Hebrew)
At a crucial time when the development and deployment of AI are rapidly evolving, experts are looking at ways we can use quantum computing to protect AI from its vulnerabilities.
Machine learning is a field of artificial intelligence where computer models become experts in various tasks by consuming large amounts of data, instead of a human explicitly programming their level of expertise. These algorithms do not need to be taught but rather learn from seeing examples, similar to how a child learns.
Just about verything will have AI in the future.
“The compact full-frame camera promises advanced autofocus features, performance, and high-quality 4K video. Following the lead of recent Sony cameras, the a7C II also includes a dedicated artificial intelligence (AI) processing engine to drive some of its more sophisticated photo and video features, including robust subject recognition, real-time tracking, and AI-based Auto Framing.”
The Sony a7C II is available body only for $2,199.99 or in a kit with Sony’s 28-60mm zoom lens for $2,499.99. This is a $300 difference compared to the Sony a7 IV body. The Sony a7C II is available in gray and black colorways. The Sony a7C II will begin shipping this fall.
Sony a7CR: a7R V Sensor in a Compact Body.
Alongside the Sony a7C II, Sony has announced its first a7CR camera. Like the a7R series, including its most recent iteration, the Sony a7R V, the Sony a7CR offers a high-resolution image sensor, albeit in a compact package.

Experts warn that AI-generated content may pose a threat to the AI technology that produced it.
In a recent paper on how generative AI tools like ChatGPT are trained, a team of AI researchers from schools like the University of Oxford and the University of Cambridge found that the large language models behind the technology may potentially be trained on other AI-generated content as it continues to spread in droves across the internet — a phenomenon they coined as “model collapse.” In turn, the researchers claim that generative AI tools may respond to user queries with lower-quality outputs, as their models become more widely trained on “synthetic data” instead of the human-made content that make their responses unique.
Other AI researchers have coined their own terms to describe the training method. In a paper released in July, researchers from Stanford and Rice universities called this phenomenon the “Model Autography Disorder,” in which the “self-consuming” loop of AI training itself on content generated by other AI could result in generative AI tools “doomed” to have their “quality” and “diversity” of images and text generated falter. Jathan Sadowski, a senior fellow at the Emerging Technologies Research Lab in Australia who researches AI, called this phenomenon “Habsburg AI,” arguing that AI systems heavily trained on outputs of other generative AI tools can create “inbred mutant” responses that contain “exaggerated, grotesque features.”
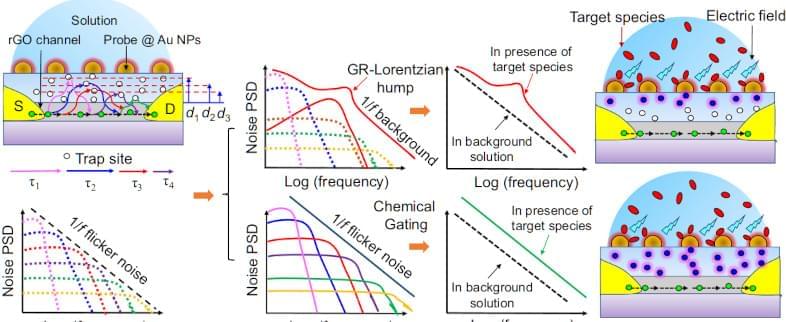
If you live in a place where you can buy Arduinos and Raspberry Pis locally, you probably don’t spend much time worrying about your water supply. But in some parts of the world, it is nothing to take for granted, bad water accounts for as many as 500,000 deaths worldwide every year. Scientists have reported a graphene sensor they say costs a buck and can detect dangerous bacteria and heavy metals in drinking water.
The sensor uses a GFET — a graphene-based field effect transistor to detect lead, mercury, and E. coli bacteria. Interestingly, the FETs transfer characteristic changes based on what is is exposed to. We were, frankly, a bit surprised that this is repeatable enough to give you useful data. But apparently, it is especially when you use a neural network to interpret the results.
What’s more, there is the possibility the device could find other contaminants like pesticides. While the materials in the sensor might have cost a dollar, it sounds like you’d need a big equipment budget to reproduce these. There are silicon wafers, spin coating, oxygen plasma, and lithography. Not something you’ll whip up in the garage this weekend.

Cell therapy company BlueRock Therapeutics has revealed promising results from the Phase 1 clinical trial of bemdaneprocel, an investigational stem cell therapy designed to treat Parkinson’s disease. BlueRock, a wholly owned independently operated subsidiary of pharma giant Bayer, is creating a pipeline of cell therapy treatments for neurological, cardiovascular, immunological, and ophthalmic conditions.
Bemdaneprocel is a stem cell therapy developed to replace dopamine-producing neurons that are lost in Parkinson’s disease. Derived from pluripotent stem cells, the therapy involves implanting neuron precursors into the brains of patients to potentially restore neural networks and improve motor and non-motor functions.
“The need for new therapies to help patients struggling with Parkinson’s disease is clear,” said Ahmed Enayetallah, Head of Development at BlueRock Therapeutics. “We are excited to be sharing the results of this Phase 1 and look forward to advancing bemdaneprocel to the next stage of clinical testing.”
Quantum computing is on the verge of catapulting the digital revolution to new heights.
Turbocharged processing holds the promise of instantaneously diagnosing health ailments and providing rapid development of new medicines; greatly speeding up response time in AI systems for such time-sensitive operations as autonomous driving and space travel; optimizing traffic control in congested cities; helping aircraft better navigate extreme turbulence; speeding up weather forecasting that better prepares localities facing potential disaster, and optimizing supply chain systems for more efficient delivery times and cost savings.
But we’re not there yet. One of the greatest obstacles facing quantum operations is error-correction.

The recent surge in generative artificial intelligence (AI) technology is boosting the tech industry after growth slowed across the sector earlier this year.
As federal interest rates rose and the tech industry was coming off a pandemic-induced high, the once-vibrant sector suffered through lower earnings and layoffs throughout the year.

Now, you can use Google’s AI to make spreadsheets, whip up slide decks, and summarize all those documents you were never going to actually read.
Google announced today that it is rolling out its Duet AI assistant across all of its Workspace apps, including Gmail, Drive, Slides, Docs, and more. The Duet tech has been in testing for a while, the company said, with more than a million people already kicking the tires on Google’s virtual assistant. Now, it’s coming to anyone paying for Google’s Workspace apps.
Google announced Duet AI at its I/O developer conference earlier this year, pitching the collection of features as a helpful collaborator in all your Google apps. You might ask Duet to turn… More.
AI is coming to help you make spreadsheets, read long docs, and look better in meetings.
How will AI affect businesses and employees? It’s the million-dollar question, and according to Harvard Business School’s Raffaella Sadun, the answer will depend on how well an organization connects the new technologies to both a broad corporate vision and individual employee growth.
One without the other is a recipe for job elimination and fewer new opportunities for all. Luckily, she points out, we are early in our AI journey, and nothing is predetermined. Smart leaders don’t need to understand every technicality of AI. But they do need to identify the best use cases for their specific business and communicate a clear strategy for reskilling their teams.
For this episode of our video series “The New World of Work”, HBR editor in chief Adi Ignatius sat down with Sadun, who wrote the HBR article, “Reskilling in the Age of AI” (https://hbr.org/2023/09/reskilling-in-the-age-of-ai), to discuss:
• How leaders should use GenAI to augment their own decision making, without entrusting it to make the actual decisions.
• Even in the age of AI, the top management skills will be a mixture of technical (“hard”) and social (“soft”) skills. Those who excel will comprehend their organization’s complexity while communicating a clear vision to all employees.
• Handling change management when everyone is uncertain about the future and regular employees are especially fearful.
This interview part of a series called “The New World of Work,” which explores how top-tier executives see the future and how their companies are trying to set themselves up for success. Each week, Adi will interview a leader on LinkedIn Live — and then share an inside look at those conversations and solicit questions for future discussions in a newsletter just for HBR subscribers. If you’re a subscriber, you can sign up for the newsletter here: https://hbr.org/my-library/preferences?movetile=newworldofwork.
Follow us: