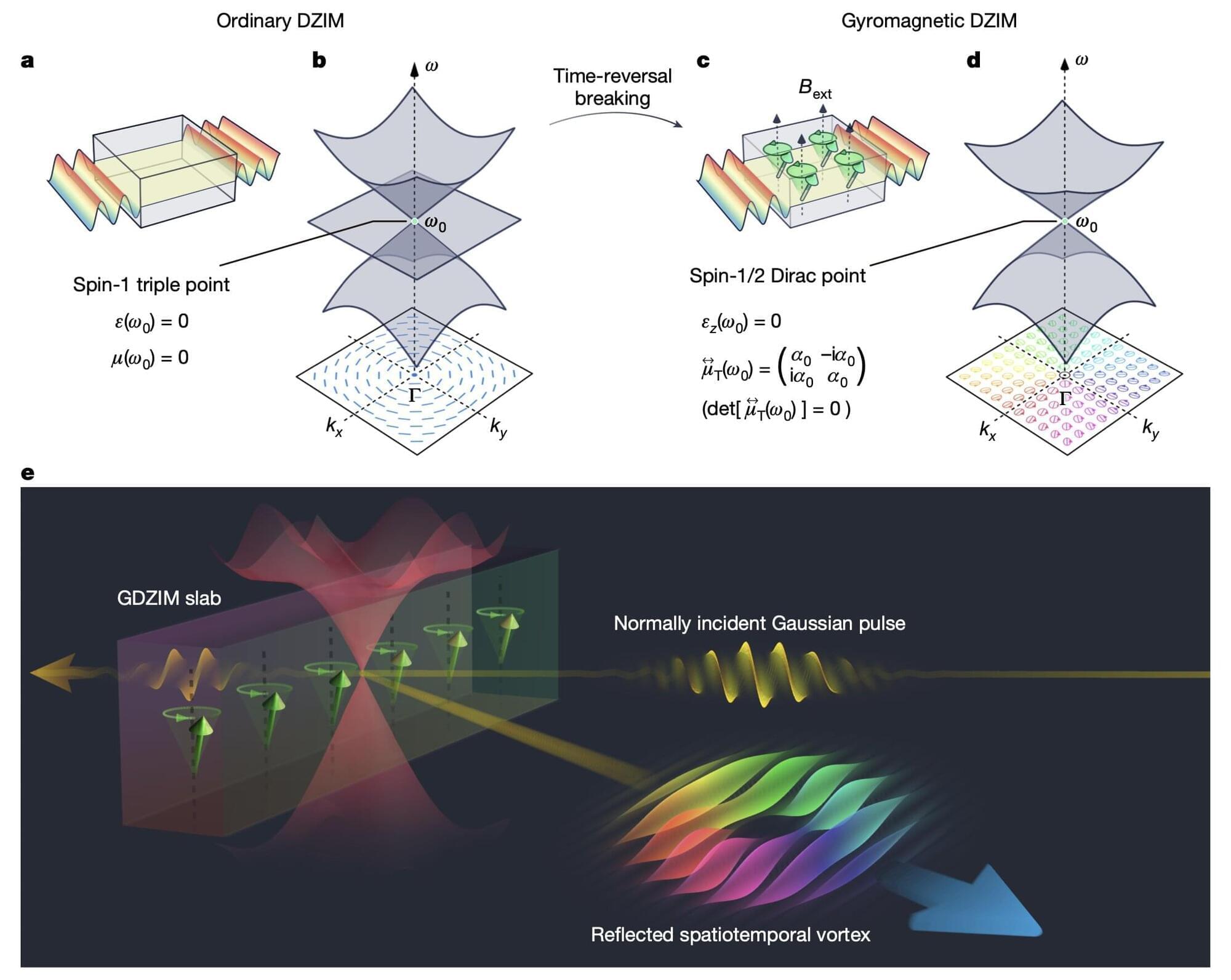
The Hong Kong University of Science and Technology (HKUST)-led research team has adopted gyromagnetic double-zero-index metamaterials (GDZIMs) — a new optical extreme-parameter material – and developed a groundbreaking method to control light using GDZIMs. This discovery could revolutionize fields like optical communications, biomedical imaging, and nanotechnology, enabling advances in integrated photonic chips, high-fidelity optical communication, and quantum light sources.
Published in Nature, the study was co-led by Prof. CHAN Che-Ting, Interim Director of the HKUST Jockey Club Institute for Advanced Study and Chair Professor in the Department of Physics, and Dr. ZHANG Ruoyang, Visiting Scholar in the Department of Physics at HKUST.