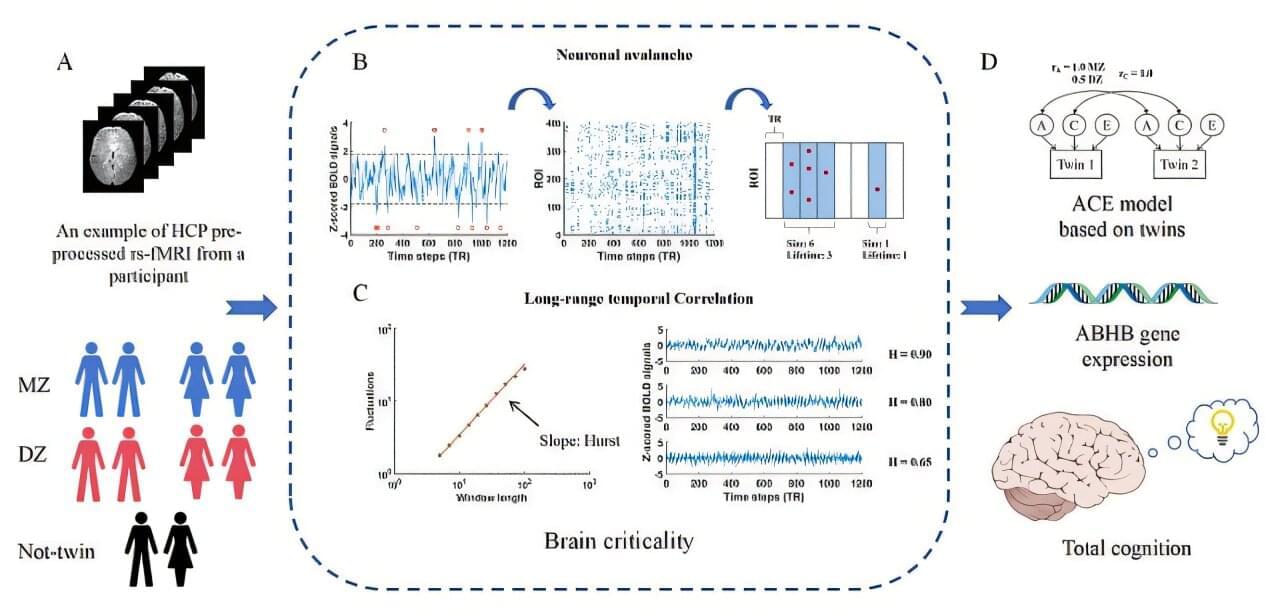
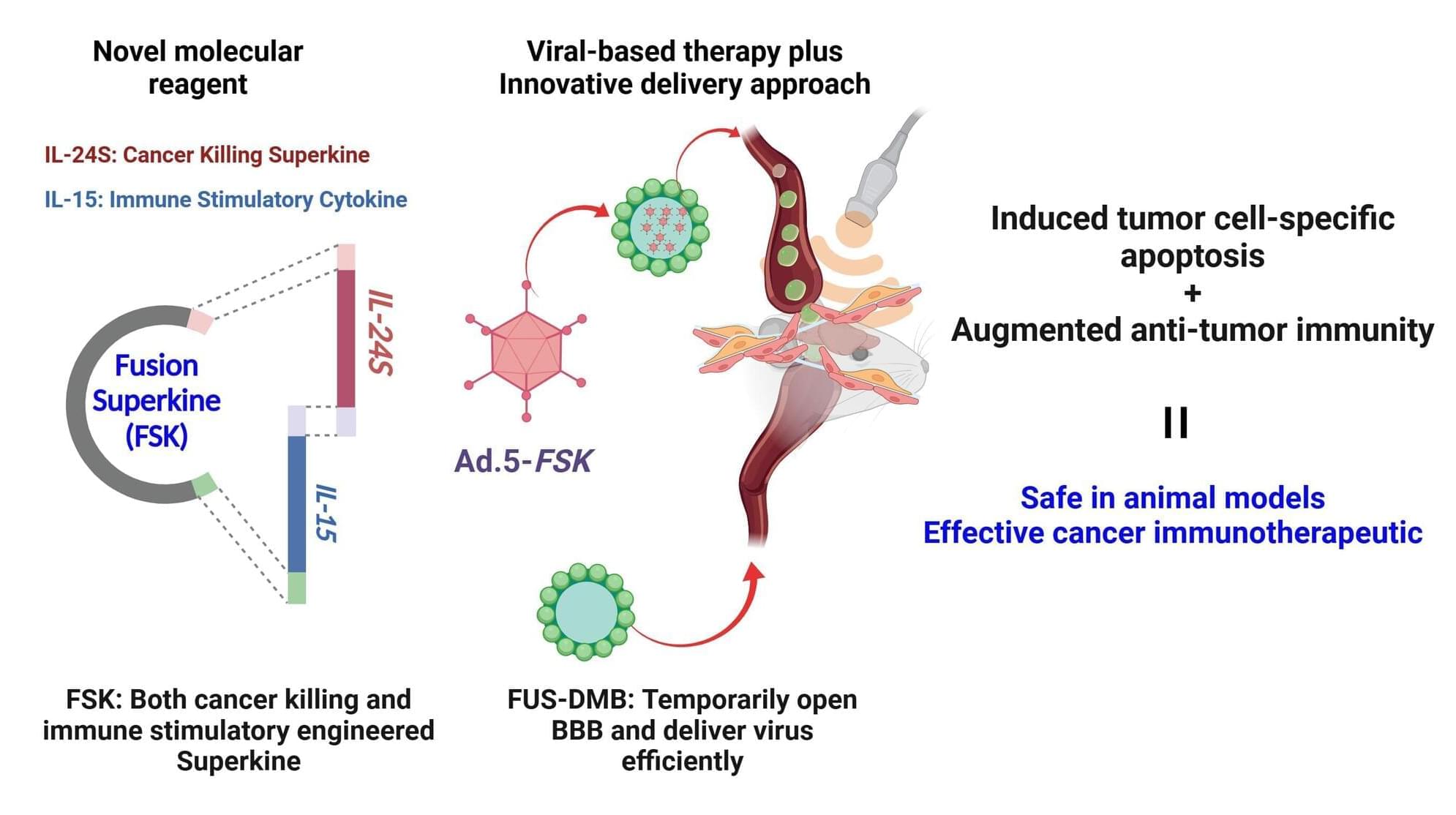
An MRI scan revealed a brain tumor located in a difficult area, and performing a biopsy would carry significant risks for the patient, who had initially sought medical help due to double vision. Cases like this, discussed by a multidisciplinary team of cancer specialists, led researchers at Charité – Universitätsmedizin Berlin, along with their collaborators, to search for alternative diagnostic methods.
Their solution is an AI model that analyzes specific features in the genetic material of tumors, particularly their epigenetic fingerprint, which can be obtained from sources such as cerebrospinal fluid. As reported in the journal Nature Cancer, the model classifies tumors both rapidly and with high accuracy.