She worked in top labs at Stanford and Harvard. Now she wants to disrupt a 100-Billion-Dollar market by rejuvenating your skin using genetic engineering.


The lone volunteer in a unique study involving a gene-editing technique has died, and those behind the trial are now trying to figure out what killed him.
Terry Horgan, a 27-year-old who had Duchenne muscular dystrophy, died last month, according to Cure Rare Disease, a Connecticut-based nonprofit founded by his brother, Rich, to try and save him from the fatal condition.
Although little is known about how he died, his death occurred during one of the first studies to test a gene editing treatment built for one person. It’s raising questions about the overall prospect of such therapies, which have buoyed hopes among many families facing rare and devastating diseases.


With inherited gene mutations from both parents, a woman in Spain is battling with 12 tumors in her body.
As stated by the Spanish National Cancer Research Centre (CNIO), the woman first developed a tumor when still a baby and other tumors followed it within five years. 36 year-old-patient has developed twelve tumors, at least five of them malignant in her life. Each one has been of a unique kind and has affected a different area of the body.
“We still don’t understand how this individual could have formed during the embryonic stage, nor could have overcome all these pathologies,” says Marcos Malumbres, director of the Cell Division and Cancer Group at the Spanish National Cancer Research Centre (CNIO).
Genetic engineering is a rapidly progressing scientific discipline, with tremendous current application and future potential. It’s a bit dizzying for a science communicator who is not directly involved in genetics research to keep up. I do have some graduate level training in genetics so at least I understand the language enough to try to translate the latest research for a general audience.
Many readers have by now heard of CRISPR – a powerful method of altering or silencing genes that brings down the cost and complexity so that almost any genetics lab can use this technique. CRISPR is actually just the latest of several powerful gene-altering techniques, such as TALEN. CRISPR is essentially a way to target a specific sequence of the DNA, and then deliver a package which does something, like splice the DNA. But you also need to target the correct cells. In a petri dish, this is simple. But in living organism, this is a huge challenge. We have developed several viral vectors that can be targeted to specific cell types in order to deliver the CRIPR (or TALEN), which then targets the specific DNA.
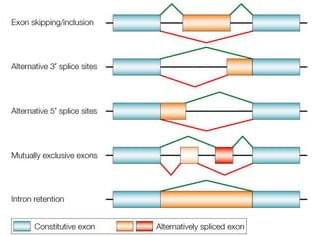
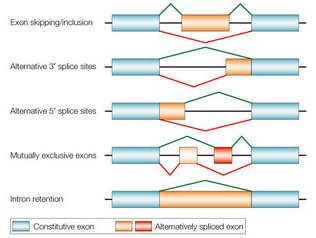
Now I would like to present a different technique I have not previously written about here – alternative splicing. A recent study presents what seems like a significant advance in this technology, so it’s a good time to review it. “Alternative splicing” refers to a natural phenomenon of genetics. Genes are composed of introns and exons. I always thought the nomenclature was counterintuitive, but the exons are actually the part of the gene that gets expressed into a protein. The introns are the part that is not expressed, so they are cut out of the gene when it is being converted into mRNA, and the exons are stitched together to form the sequence that is translated into a protein. Alternative splicing refers to the fact that the way in which the introns are removed and the exons stitched together can vary, creating alternative forms of the resulting protein.
A Ludwig Cancer Research study has developed a novel nanotechnology that triggers potent therapeutic anti-tumor immune responses and demonstrated its efficacy in mouse models of multiple cancers. Led by Co-director Ralph Weichselbaum, investigator Wenbin Lin and postdoctoral researcher Kaiting Yang at the Ludwig Center at Chicago, the study describes the synthesis, mechanism of action and preclinical assessment of the nanoparticle, which is loaded with a drug that activates a protein central to the efficient induction of anti-cancer immunity. The study, which overcomes significant technical barriers to targeting that protein-;stimulator of interferon genes, or STING-;for cancer therapy, appears in the current issue of Nature Nanotechnology.
“The nanoparticles developed by the Lin lab release a drug that targets macrophages and can activate potent antitumor immune responses that extend the survival of mice bearing a variety of tumors,” said Chicago Center Co-director Weichselbaum. “When used in combination with radiotherapy and immunotherapy, they even help control ‘cold tumors’ that are otherwise almost completely impervious to immune attack.”
STING is part of cellular sensing system for DNA fragments, which are generated by infection or cancer treatments that damage DNA, like radiotherapy and some chemotherapies. Its activation promotes inflammation and drives immune cells like macrophages and dendritic cells to process and present cancer antigens to T cells, helping to stimulate and direct the immune assault on tumors. Though STING is a prized target for drug development, the drug-like molecules that can activate the molecular sensor-;known as cyclic dinucleotides (CDNs)-;have been plagued by issues of poor bioavailability, low stability and high toxicity in the absence of any means to target them specifically to tumors.
Circa 1999 this can lead to genetic editing that allows people to handle even a nuclear fallout level of radiation and even allow them to handle outer space better.
Rockville, MD — No, it’s not the cockroach, but rather a strain of pink bacteria that can survive 1.5 million rads of gamma irradiation — a dose 3,000 times the amount that would kill a human. This dose of radiation shreds the bacteria’s genome into hundreds of pieces. The organism’s remarkable ability to repair this DNA damage completely in a day and go on living offers researchers tantalizing clues to better understanding the mechanism of cellular repair. Advances in this area could in turn improve our understanding of cancer which is frequently caused by unrepaired DNA damage. Genetically engineering the microbe could lead to improved ways to cleanup pollution and to new industrial processes.
U.S. Department of Energy-funded researchers at The Institute for Genomic Research (TIGR) describe the complete genetic sequence of the bacteria Deinococcus radiodurans in the November 19 issue of Science.
“This is a significant accomplishment,” Secretary of Energy Bill Richardson said. “The Department of Energy began microbial genome work to support bold science and to help meet our unique environment and energy mission needs. Besides the insights into the way cells work, this new research may help provide a new safe and inexpensive tool for some of the nation’s most difficult cleanup challenges.”

Researchers examining post-mortem brains confirm a long-held hypothesis explaining neurotransmitter’s connection to a debilitating disorder.
How does the brain chemical dopamine relate to schizophrenia? It is a question that vexed scientists for more than 70 years, and now researchers at the Lieber Institute for Brain Development (LIBD) believe they have solved the challenging riddle. This new understanding may lead to better treatment of schizophrenia, an often-devastating brain disorder characterized by delusional thinking, hallucinations, and other forms of psychosis.
Through their exploration of the expression of genes in the caudate nucleus – a region of the brain linked to emotional decision-making – the scientists uncovered physical evidence that neuronal cells are unable to precisely control levels of dopamine. They also identified the genetic mechanism that controls dopamine flow. Their findings were published today (November 1) in the journal Nature Neuroscience.
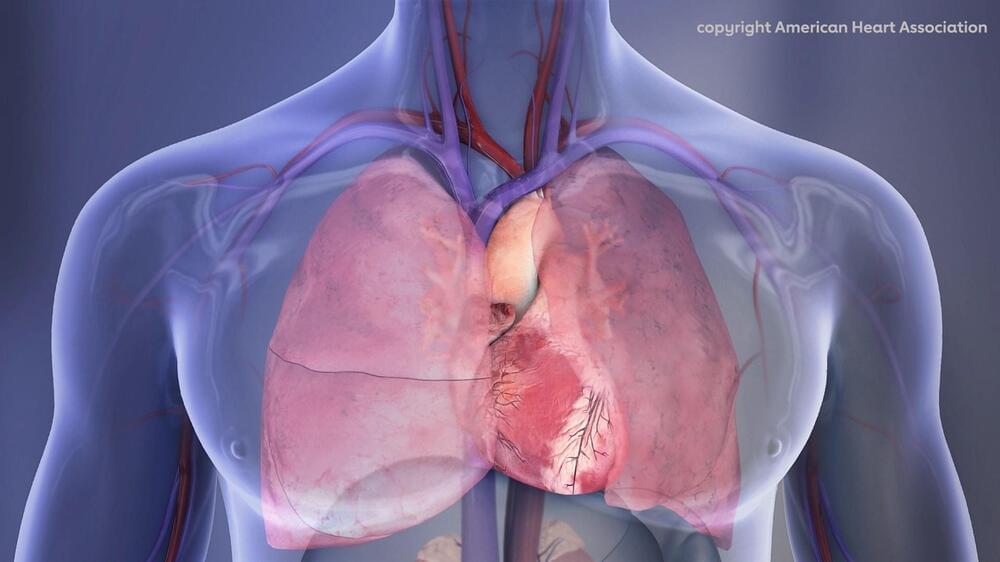
A recent study to be presented at the American Heart Association (AHA) Scientific Sessions 2022 revealed unexpected changes in the electrical conduction system of the first genetically-modified porcine-to-human heart xenotransplant.
Xenotransplantation is the procedure of transplantation/implantation into a human of organs from non-human animal sources. The first pig-to-human heart xenograft was transplanted in January 2022 at the University of Maryland. The recipient survived for 61 days after receiving the xenograft. Research efforts have been underway for this xenotransplantation for over three decades.
Harvesting genetically-modified porcine hearts, the genes of which have been altered for safe transplantation into humans, would become a reality if successful. However, xenotransplantation of organs into a human carries several inherent challenges. With these transplant procedures, there is always the risk of graft rejection, infection, and abnormal heart rhythms.
(https://hannalabweb.weizmann.ac.il/) is a Senior Scientist and Professor in the Department of Molecular Genetics at the Weizmann Institute of Science in Israel, where his lab, and the interdisciplinary group of scientists within it, are focused on understanding the complexity of early embryonic stem cell biology and early developmental dynamics, as well as advancing human disease modeling.
More specifically, Dr. Hanna’s lab investigates the detailed process of cellular reprogramming, in which induced pluripotent stem cells are generated from somatic cells, and they investigate how pluripotency is maintained throughout development in mouse and human. In their studies they employ a diverse arsenal of biological experimentation methods, high throughput screening, advanced microscopy and genomic analyses seeking to combine biological experimentation with computational biology, theory and modeling, to elucidate various biological questions.
Dr. Hanna completed both his MD and PhD at The Hebrew University of Jerusalem, where his work was focused in the domain of immunology with his thesis focus on novel molecular and functional properties of human NK Subsets. He then went on to do postdoctoral studies at the Whitehead Institute for Biomedical Research at MIT under the tutelage of Prof. Dr. Rudolf Jaenisch with a field of study of pluripotency and epigenetic reprogramming.
Dr. Hanna is also Founder and Chief Scientific Advisor, of Renewal Bio (https://www.renewal.bio/), a biotech company looking to leverage the power of these new stem cell technologies, potentially applying them to a wide variety of human ailments including infertility, genetic diseases, and longevity.