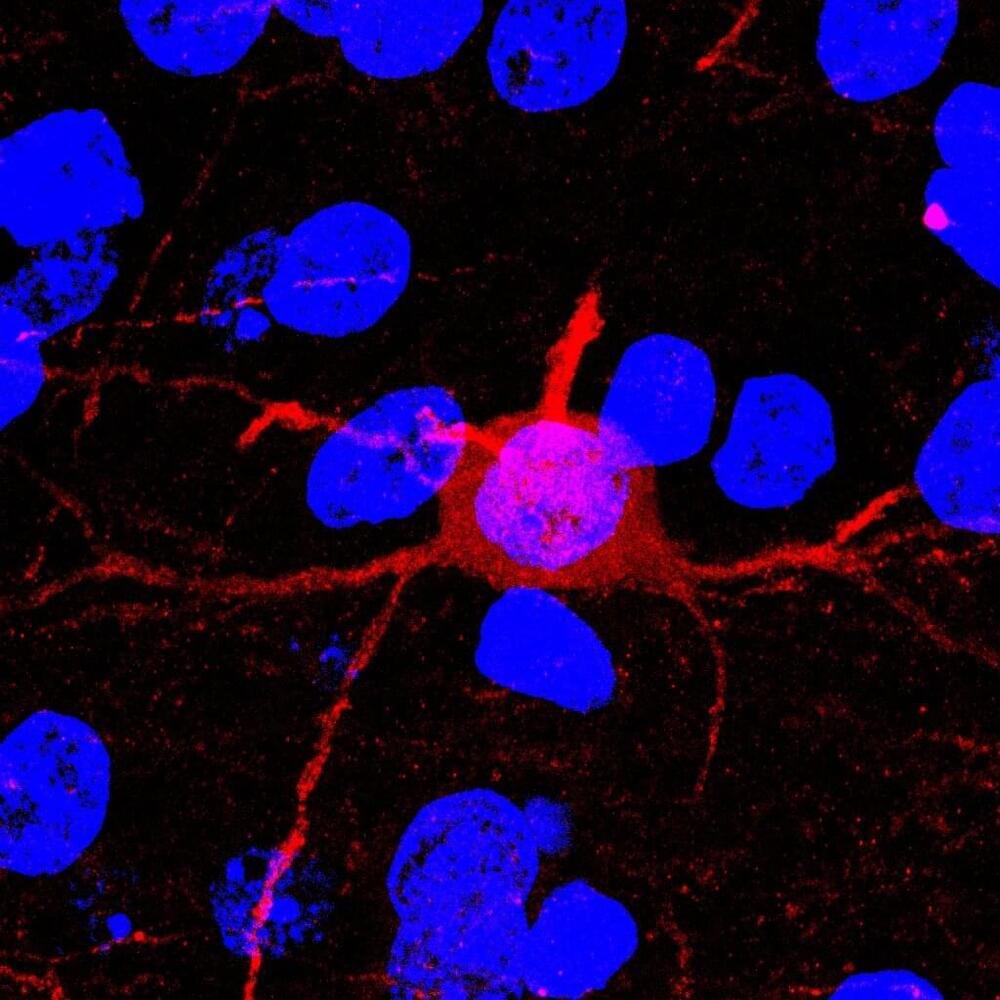
Substance use disorder (SUD) is an extremely difficult disorder to overcome, and many individuals with SUD return to regular use after repeated attempts to quit.
A return to regular drug use can be caused by the body’s physical dependence on the drug as well as experiences associated with prior drug use. Exactly how these drug associations are formed in the brain and how they trigger a return to drug use remain unclear.
“Individuals make long-lasting associations between the euphoric experience of the drug and the people, places and things associated with drug use,” said Christopher Cowan, Ph.D. professor in the Department of Neuroscience at the Medical University of South Carolina (MUSC) and member of the Brain and Behavior Research Foundation Scientific Council.