Meet the $15 super computer that’s the size of an iPhone.
Category: computing – Page 842
Engineers develop room temperature, two-dimensional platform for quantum technology
Possible quantum computing at room temperature. Scientists working with hexagonal boron nitride, which allows them to work in two-dimensional arrays. Simpler than using 3D objects such as diamonds.
Researchers have now demonstrated a new hardware platform based on isolated electron spins in a two-dimensional material. The electrons are trapped by defects in sheets of hexagonal boron nitride, a one-atom-thick semiconductor material, and the researchers were able to optically detect the system’s quantum states.
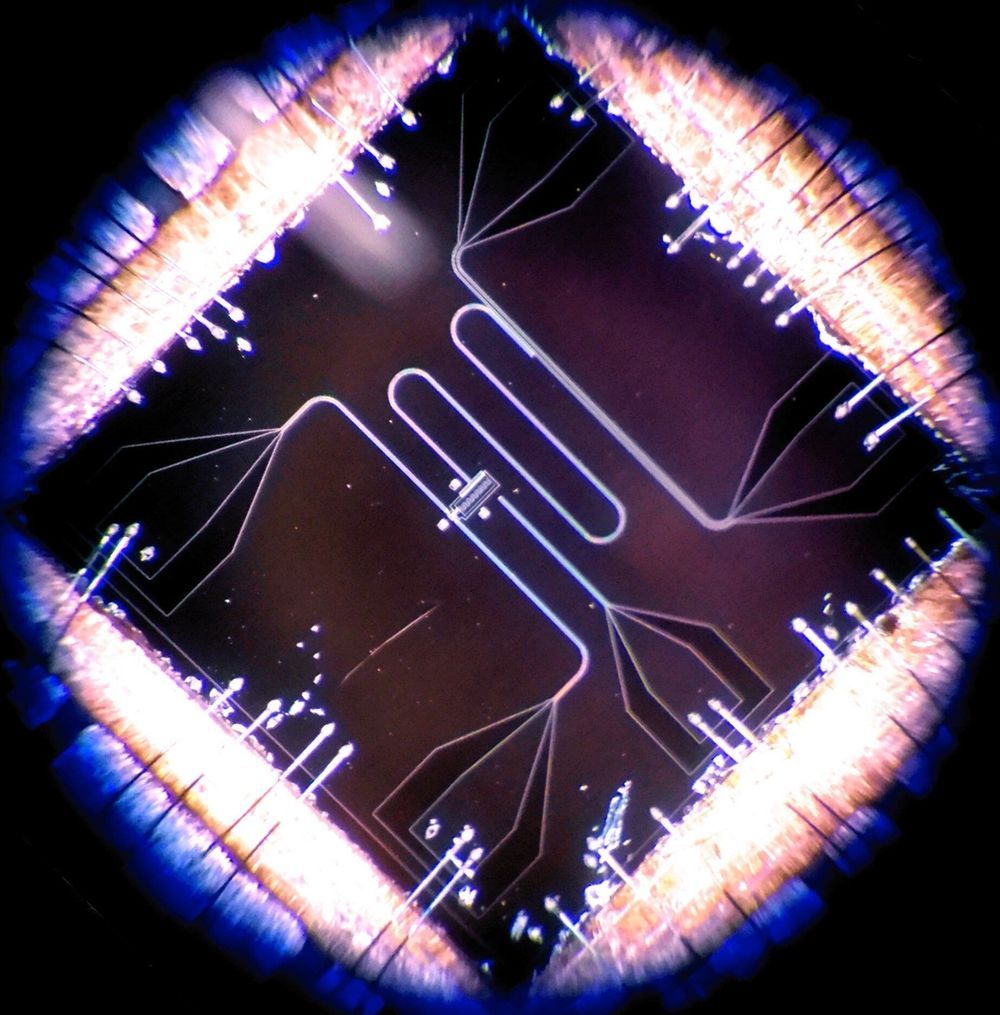
Life on the edge in the quantum world
Quantum physics sets the laws that dominate the universe at a small scale. The ability to harness quantum phenomena could lead to machines like quantum computers, which are predicted to perform certain calculations much faster than conventional computers. One major problem with building quantum processors is that the tracking and controlling quantum systems in real time is a difficult task because quantum systems are overwhelmingly fragile: Manipulating these systems carelessly introduces significant errors in the final result. New work by a team at Aalto could lead to precise quantum computers.
The researchers report controlling quantum phenomena in a custom-designed electrical circuit called a transmon. Chilling a transmon chip to within a few thousandths of a degree above absolute zero induces a quantum state, and the chip starts to behave like an artificial atom. One of the quantum features that interests researchers is that the energy of the transmon can only take specific values, called energy levels. The energy levels are like steps on a ladder: A person climbing the ladder must occupy a step, and can’t hover somewhere between two steps. Likewise, the transmon energy can only occupy the set values of the energy levels. Shining microwaves on the circuit induces the transmon to absorb the energy and climb up the rungs of the ladder.
In work published 8 February in the journal Science Advances, the group from Aalto University led by Docent Sorin Paraoanu, senior university lecturer in the Department of Applied Physics, has made the transmon jump more than one energy level in a single go. Previously, this has been possible only by very gentle and slow adjustments of the microwave signals that control the device. In the new work, an additional microwave control signal shaped in a very specific way allows a fast, precise change of the energy level. Dr. Antti Vepsäläinen, the lead author, says, “We have a saying in Finland: ‘hiljaa hyvää tulee’ (slowly does it). But we managed to show that by continuously correcting the state of the system, we can drive this process more rapidly and at high fidelity.”
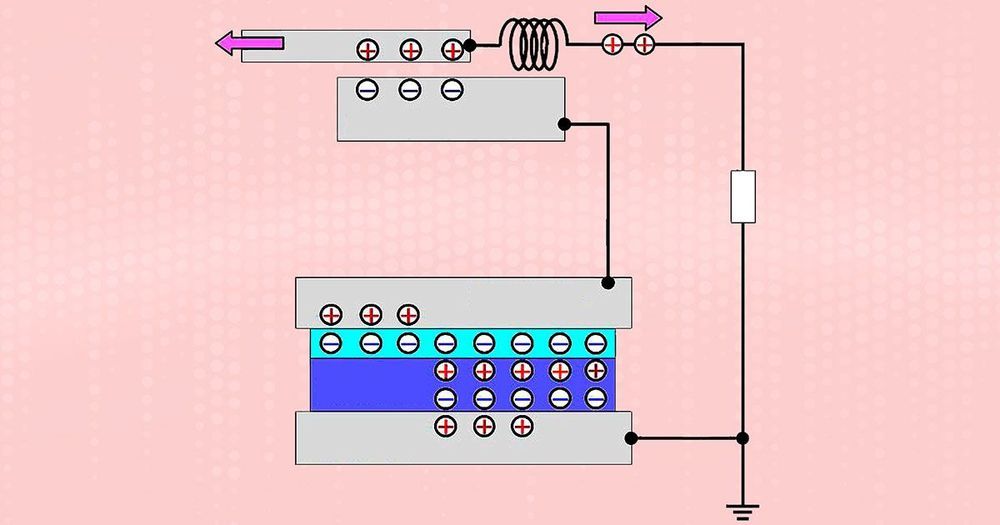
A MEMS Device Harvests Vibrations to Power the IoT
Vibration-based energy harvesting has long promised to provide perpetual power for small electronic components such as tiny sensors used in monitoring systems. If this potential can be realized, external energy sources such as batteries would no longer be needed to power these components.
Scientists at the Tokyo Institute of Technology and the University of Tokyo in Japan believe they have taken a step toward achieving self-powered components by developing a new type of micro-electromechanical system (MEMS) energy harvester. Their approach enables far more flexible designs than are currently possible— something, they say, that is crucial if such systems are to be used for the Internet of Things (IoT) and wireless sensor networks.
Scientists in Japan have developed a MEMS energy harvester charged by an off-chip electret.
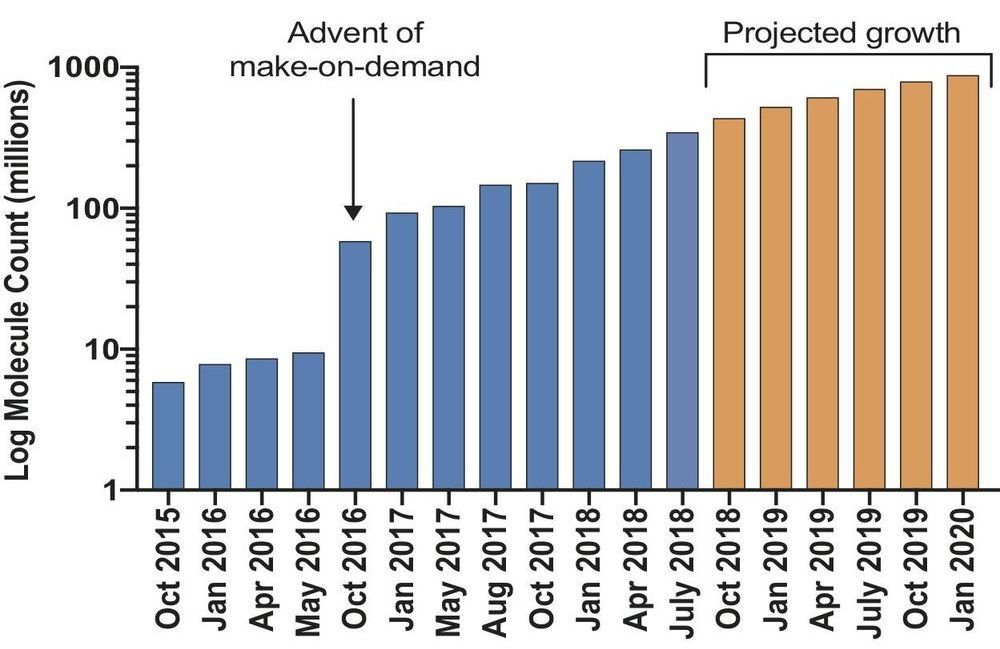
Mega docking library poised to speed drug discovery
Researchers have launched an ultra-large virtual docking library expected to grow to more than 1 billion molecules by next year. It will expand by 1000-fold the number of such “make-on-demand” compounds readily available to scientists for chemical biology and drug discovery. The larger the library, the better its odds of weeding out inactive “decoy” molecules that could otherwise lead researchers down blind alleys. The project is funded by the National Institutes of Health.
“To improve medications for mental illnesses, we need to screen huge numbers of potentially therapeutic molecules,” explained Joshua A. Gordon, M.D., Ph.D., director of NIH’s National Institute of Mental Health (NIMH), which co-funded the research. “Unbiased computational modeling allows us to do this in a computer, vastly expediting the process of discovering new treatments. It enables researchers to virtually “see” a molecule docking with its receptor protein—like a ship in its harbor berth or a key in its lock—and predict its pharmacological properties, based on how the molecular structures are predicted to interact. Only those relatively few candidate molecules that best match the target profile on the computer need to be physically made and tested in a wet lab.”
Bryan Roth, M.D., Ph.D., of the University of North Carolina (UNC) Chapel Hill, Brian Shoichet, Ph.D., and John Irwin, Ph.D., of the University of California San Francisco, and colleagues, report on their findings Feb. 6, 2019 in the journal Nature. The study was supported, in part, by grants from NIMH, National Institute of General Medical Sciences (NIGMS), the NIH Common Fund, and National Institute of Neurological Disorders and Stroke (NINDS).

HTC Vive Pro Eye Hands-On: Feeling Powerful With Built-In Tobii Eye Tracking
Hands are old news. VR navigation, control and selection is best done with the eyes—at least that’s what HTC Vive is banking on with the upcoming HTC Vive Pro Eye, a VR headset with integrated Tobii eye tracking initially targeting businesses. I tried out a beta version of the feature myself on MLB Home Run Derby VR. It’s still in development and, thus, was a little wonky, but I can’t deny its cool factor.
HTC announced the new headset Tuesday at the CES tech show in Las Vegas. The idea is that by having eye tracking built into the headset, better use cases, such as enhanced training programs, can be introduced. The VR player also says users can expect faster VR interactions and better efficiency in terms of tapping your PC’s CPU and GPU.
Of course, before my peepers could be tracked I needed to calibrate the headset for my special eyes. It was quite simple, after adjusting the interpupillary distance appropriately, the headset had me stare at a blue dot that bounced around my field of view (FOV). The whole thing took less than a minute.

Electromagnetic Pulse (EMP) Attack: A Preventable Homeland Security Catastrophe
A major threat to America has been largely ignored by those who could prevent it. An electromagnetic pulse (EMP) attack could wreak havoc on the nation’s electronic systems-shutting down power grids, sources, and supply mechanisms. An EMP attack on the United States could irreparably cripple the country. It could simultaneously inflict large-scale damage and critically limit our recovery abilities. Congress and the new Administration must recognize the significance of the EMP threat and take the necessary steps to protect against it.
Systems Gone Haywire
An EMP is a high-intensity burst of electromagnetic energy caused by the rapid acceleration of charged particles. In an attack, these particles interact and send electrical systems into chaos in three ways: First, the electromagnetic shock disrupts electronics, such as sensors, communications systems, protective systems, computers, and other similar devices. The second component has a slightly smaller range and is similar in effect to lightning. Although protective measures have long been established for lightning strikes, the potential for damage to critical infrastructure from this component exists because it rapidly follows and compounds the first component. The final component is slower than the previous two, but has a longer duration. It is a pulse that flows through electricity transmission lines-damaging distribution centers and fusing power lines. The combination of the three components can easily cause irreversible damage to many electronic systems.
New invisibility cloak hides tiny three-dimensional objects of any shape
Circa 2015
Scientists at UC Berkeley have developed a foldable, incredibly thin invisibility cloak that can wrap around microscopic objects of any shape and make them undetectable in the visible spectrum. In its current form, the technology could be useful in optical computing or in shrouding secret microelectronic components from prying eyes, but according to the researchers involved, it could also be scaled up in size with relative ease.
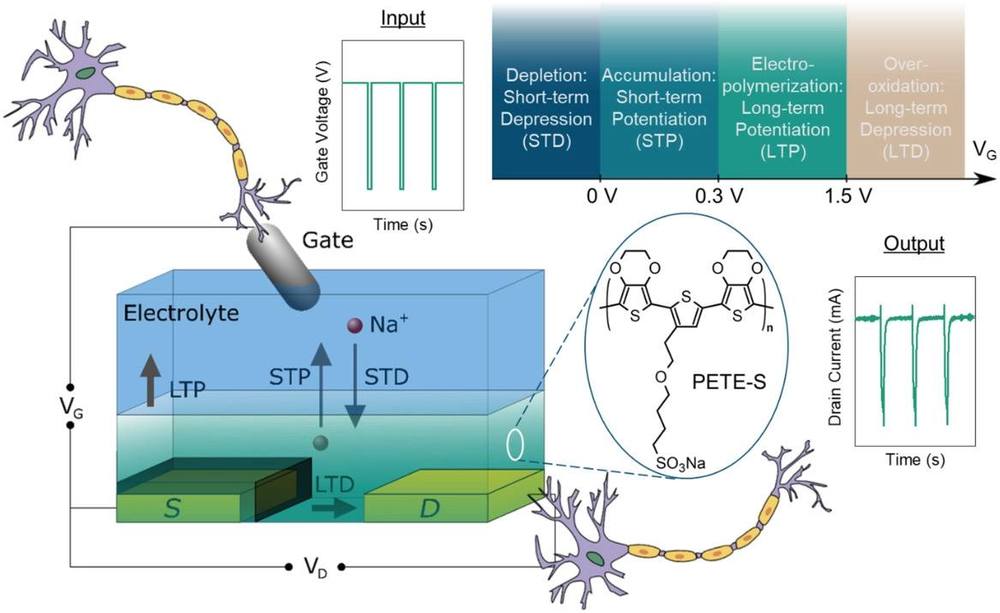
A single transistor process that can create connections
A single transistor process that can create connections, and encode short and long term memories. The transistor is based on organic versus inorganic technology. The project relies on a form of transistor that uses ion injection from an electrolyte solution, which changes the storage and connectivity of the transistor.
The end result, a simple learning circuit to replicate neuronal action.
News Article: https://www.sciencedaily.com/releases/2019/02/190205102537.htm