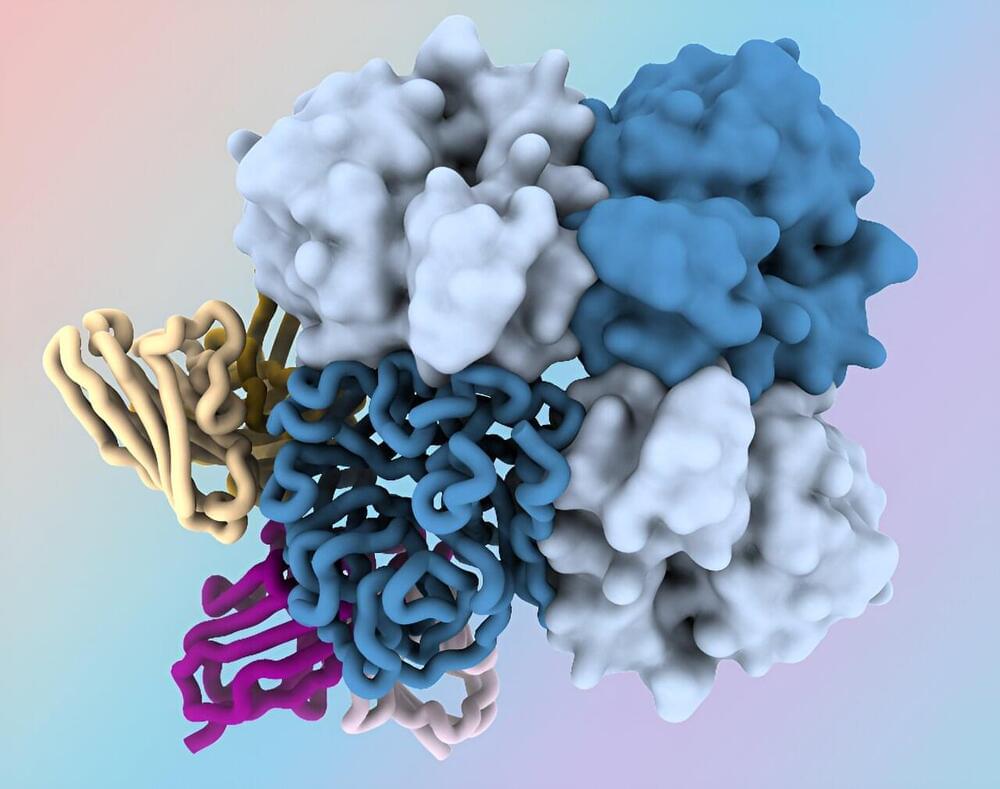
Researchers at the National Institutes of Health have identified antibodies targeting a hard-to-spot region of the influenza virus, shedding light on the relatively unexplored “dark side” of the neuraminidase (NA) protein head. The antibodies target a region of the NA protein that is common among many influenza viruses, including H3N2 subtype viruses, and could be a new target for countermeasures. The research, led by scientists at the National Institute of Allergy and Infectious Diseases’ Vaccine Research Center, part of NIH, was published today in Immunity.
Influenza, or flu, sickens millions of people across the globe each year and can lead to severe illness and death. While vaccination against influenza reduces the burden of the disease, updated vaccines are needed each season to provide protection against the many strains and subtypes of the rapidly evolving virus. Vaccines that provide protection against a broad range of influenza viruses could prevent outbreaks of new and reemerging flu viruses without the need for yearly vaccine reformulation or vaccinations.
One way to improve influenza vaccines and other countermeasures is to identify new targets on the virus’s surface proteins in “conserved” regions—portions that tend to be relatively unchanged between different strains of the virus. Influenza NA is a surface protein containing a globular head portion and a narrow stalk portion.