Figuring out how the human body can withstand underwater pressure has been a problem for over a century, but a ragtag band of divers is experimenting with hydrogen to find out.



Using the results of a standard blood test and an online tool, you can find out if you are at increased risk of having a heart attack within six months. The tool has been developed by a research group at Uppsala University in the hope of increasing patients’ motivation to change their lifestyles.
Their paper is published in the journal Nature Cardiovascular Research.
Heart attacks are the most common cause of death in the world and are increasing globally. Many high-risk people are not identified or do not take their preventive treatment.
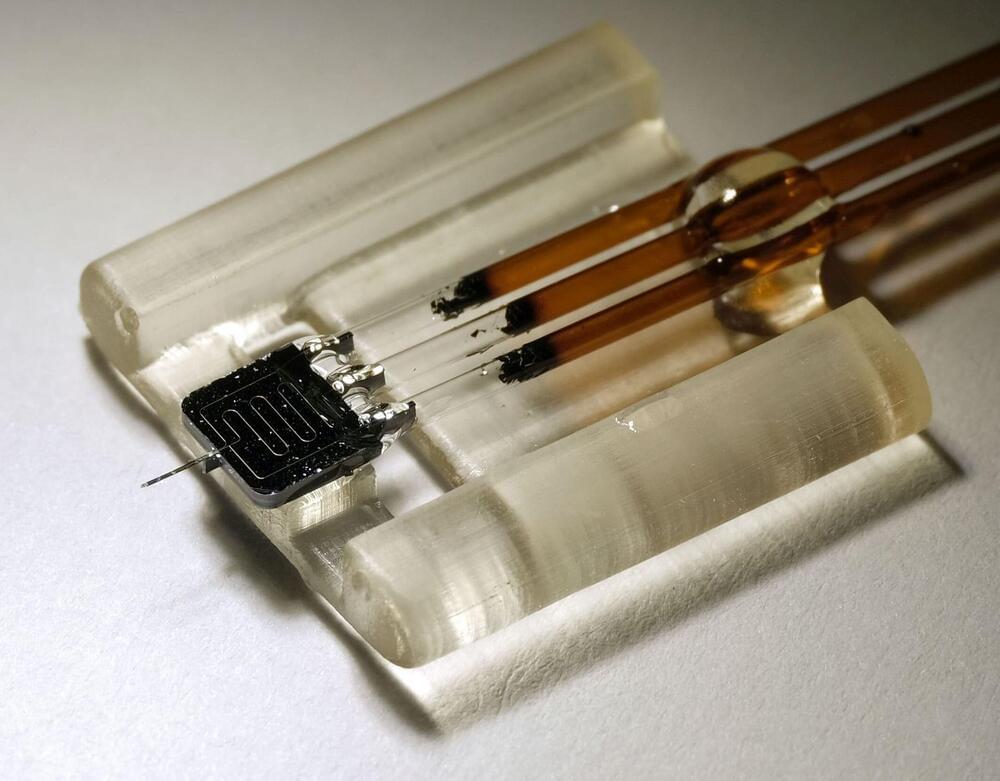
Longstanding challenges in biomedical research such as monitoring brain chemistry and tracking the spread of drugs through the body require much smaller and more precise sensors. A new nanoscale sensor that can monitor areas 1,000 times smaller than current technology and can track subtle changes in the chemical content of biological tissue with sub-second resolution, greatly outperforming standard technologies.
The device, developed by researchers at the University of Illinois Urbana-Champaign, is silicon-based and takes advantage of techniques developed for microelectronics manufacturing. The small device size enables it to collect chemical content with close to 100% efficiency from highly localized regions of tissue in a fraction of a second. The capabilities of this new nanodialysis device are reported in the journal ACS Nano.
“With our nanodialysis device, we take an established technique and push it into a new extreme, making biomedical research problems that were impossible before quite feasible now,” said Yurii Vlasov, a U. of I. electrical & computer engineering professor and a co-lead of the study. “Moreover, since our devices are made on silicon using microelectronics fabrication techniques, they can be manufactured and deployed on large scales.”


Varda plans to pioneer the use of orbital manufacturing spacecraft such as this capsule to open unique pathways for engineering materials in space. “Processing materials in microgravity, or the near-weightless conditions found in space, offers a unique environment not available through terrestrial processing,” the company’s website states.
Related: Private Varda Space capsule returns to Earth with space-grown antiviral drug aboard
The recovery made Varda only the third private company to recover an intact spacecraft from orbit, after SpaceX and Boeing.
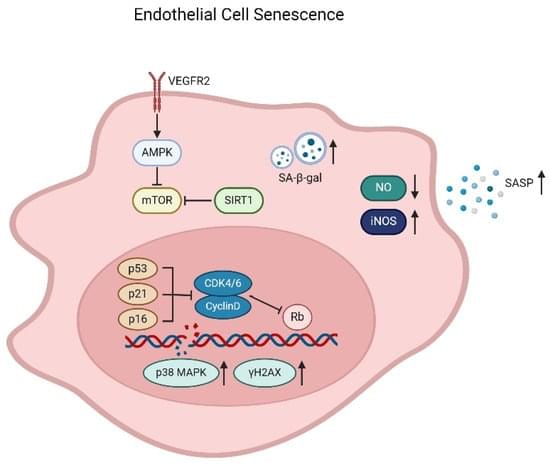
Endothelial cells line at the most inner layer of blood vessels. They act to control hemostasis, arterial tone/reactivity, wound healing, tissue oxygen, and nutrient supply. With age, endothelial cells become senescent, characterized by reduced regeneration capacity, inflammation, and abnormal secretory profile. Endothelial senescence represents one of the earliest features of arterial ageing and contributes to many age-related diseases. Compared to those in arteries and veins, endothelial cells of the microcirculation exhibit a greater extent of heterogeneity. Microcirculatory endothelial senescence leads to a declined capillary density, reduced angiogenic potentials, decreased blood flow, impaired barrier properties, and hypoperfusion in a tissue or organ-dependent manner.
A genetic marker linked to premature aging was reversed in children with obesity during a six-month diet and exercise program, according to a recent study led by the Stanford School of Medicine.
Children’s telomeres — protective molecular “caps” on the chromosomes — were longer during the weight management program, then were shorter again in the year after the program ended, the study found. The research was published last month in Pediatric Obesity.
Like the solid segment at the end of a shoelace, telomeres protect the ends of chromosomes from fraying. In all people, telomeres gradually shorten with aging. Various conditions, including obesity, cause premature shortening of the telomeres.
Autoimmune disease occurs from the body’s immune system attacking its healthy cells. Unfortunately, the mechanism that would normally prevent autoimmunity is not present in some individuals. T cells are the immune cell population responsible for killing or lysing invading pathogens. In the context of autoimmunity, T cells attack and lyse healthy cells. The thymus gland educates or prepares T cells to become activated and target foreign pathogens. T cells are exposed to different molecules and surface markers which further train these cells on how to respond when they come into contact with foreign markers. Autoimmune disorders are rare and can often be detected in children. However, there are limited treatment options, and a cure has not been found. Researchers are currently working to better treat autoimmune disorders and improve the quality of life in patients.
A recent article published in Nature, by a team led by Dr. Thomas Korn, reported a previously unknown mechanism underlying autoimmune disease. Korn is a Professor of Experimental Nueroimmunology at the Technical University of Munich (TUM) and Principal Investigator at the Maximilian University of Munich (LMU). His lab focuses on T cell biology and the underlying mechanisms of autoimmune disorders. Korn and others demonstrated that another immune cell population, B cells, aid in T cell education in the thymus gland. Korn and others point out that B cells are part of T cell development and play a critical role in autoimmune disorder.
Researchers used both animal models and human tissue samples to conduct their research to investigate T cell development. The autoimmune disorder Korn and his team used as a model is known as neuromyelitis optica, which is similar to multiple sclerosis (MS). Researchers chose this specific model due to the well-known fact that T cells respond to the protein AQP4 in this autoimmune disorder. Interestingly, AQP4 is highly expressed in the nervous system, which becomes the target of autoimmunity. Researchers discovered that B cells also express AQP4, which present this protein to the T cells in the thymus. Interestingly, if the B cells did not express AQP4, then T cells would not become reactive to the surface protein and target healthy nervous system cells. Epithelial cells also expressed the AQP4 protein and resulted in the same autoimmune reaction. However, B cells were found to significantly impact T cell development compared to other cells in the thymus.
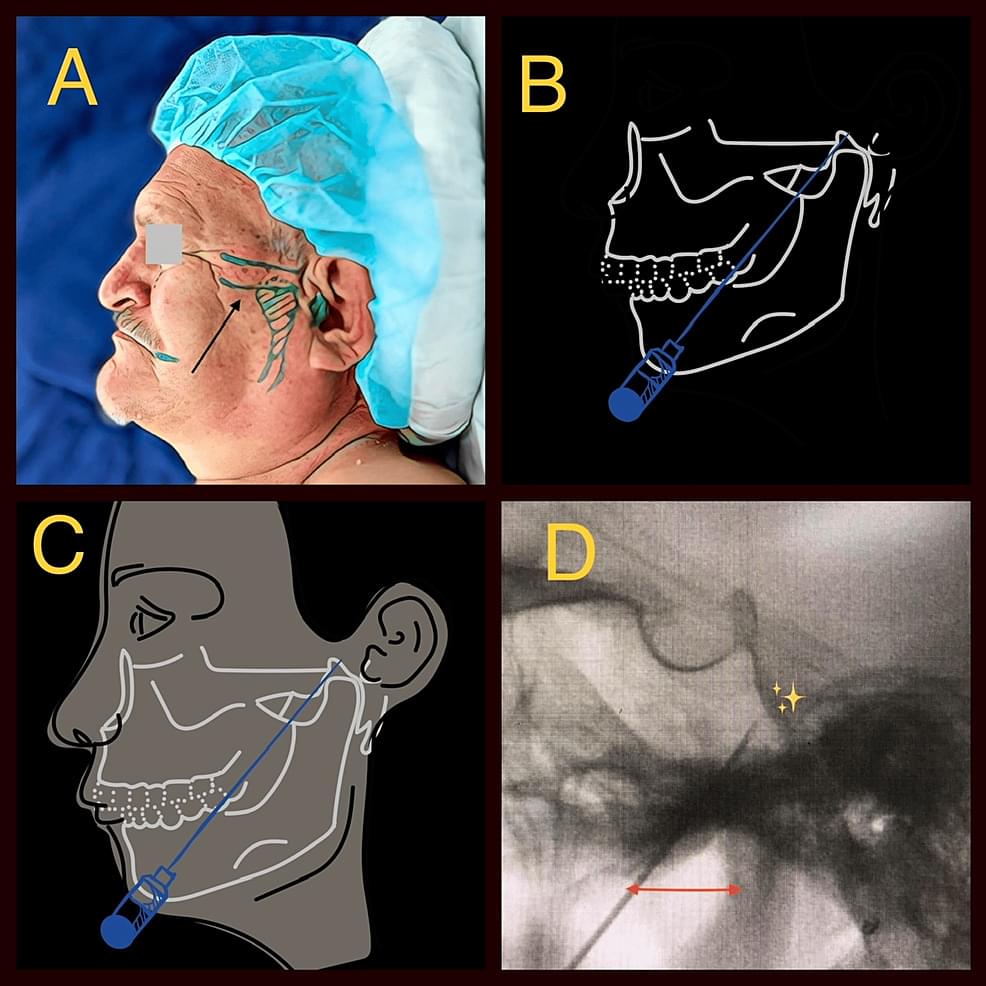
Objectives: Trigeminal neuralgia (TN) represents one of the most powerful manifestations of neuropathic pain. The diagnostic criteria, as well as its therapeutic modalities, stand firmly established. The percutaneous radiofrequency thermorhizotomy of the gasserian ganglion and posterior root of the trigeminal nerve stands as a widely employed procedure in this context. In this retrospective observational investigation, we undertake a comparative analysis of patients subjected to treatment employing continuous radiofrequency (C-rF) versus pulsed radiofrequency (P-rF).
Materials and methods: A cohort of 128 patients afflicted with essential neuralgia of the trigeminal nerve, all under the care of the distinguished author (JCA), underwent percutaneous radiofrequency thermorhizotomy between the years 2005 and 2022. They were stratified into two cohorts: Group 1 encompassed 76 patients treated with C-rF, while Group 2 comprised 52 patients subjected to P-rF intervention. All participants met the stringent inclusion and exclusion criteria for TN, with a notable concentration in the V2 and V3 territories accounting for 60% and 45%, respectively. The post-procedural follow-up period exhibited uniformity, spanning from six months to 16 years. Preceding the intervention, all patients uniformly reported a visual analog scale (VAS) score surpassing 6/10. Additionally, everyone had been undergoing pharmacological management, involving a combination of antineuropathic agents and low-potency opioids.
Results: The evaluation of clinical improvement was conducted across three temporal domains: the immediate short-term (less than 30 days), the intermediate-term (less than one year), and the prolonged-term (exceeding one year). In the short term, a noteworthy alleviation of pain, surpassing the 50% threshold, was evident in most patients (94%), a similarity observed in both cohorts (98% in Group 1 and 90% in Group 2). The VAS revealed an average rating of 3/10 for Group 1 and 2/10 for Group 2. Moving to the intermediate term, more than 50% improvement in pain was registered in 89% of patients (92% in Group 1 and 86% in Group 2). The mean VAS score stood at 3.5÷10, marginally higher in Group 2 at 4/10 compared to 3/10 in Group 1. In the final assessment, a 50% or greater reduction in pain was reported by 75% of patients, with no discernible disparity between the two cohorts. Among the cohort, 18 individuals necessitated a subsequent percutaneous intervention (10 in Group 1 and eight in Group 2), while microvascular decompression was performed on six patients (equitably distributed between the two groups), and radiosurgery was administered to three patients in Group 1.