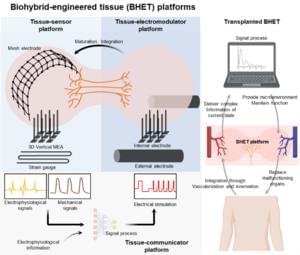
On the morning of April 26, Tsinghua University held an inauguration ceremony for Tsinghua AI Agent Hospital and the 2025 Tsinghua Medicine Townhall Meeting at the Main Building Reception Hall. Tsinghua President Li Luming and Vice President Wang Hongwei attended the event.
President Li Luming reviewed the progress of Tsinghua University’s medical programs over the past year, emphasizing the University’s strong commitment to the development of medical disciplines. He highlighted Tsinghua’s strength in fundamental research in Artificial intelligence, which has already led to a series of high-level innovations at the intersection of AI and medicine. The establishment of the Tsinghua AI Agent Hospital represents a new initiative by Tsinghua to leverage its strengths in science and engineering to empower the advancement of medicine.
President Li encouraged Tsinghua Medicine to remain committed to fostering virtue and talent, cultivating a new generation of medical innovators with both a strong medical foundation and AI literacy. He also called for deeper integration across disciplines, particularly between engineering and medicine, as well as closer ties between clinical practice and technology. Finally, he urged Tsinghua Medicine to align its work with cutting-edge global trends and national strategic needs, driving medical advancement and contributing to the protection of public health.