Finding early signs of dementia in the back of the eye may be a way to catch the disease early and start preventive treatment, a study says.

Michael Levin is a biologist at Tufts University working on novel ways to understand and control complex pattern formation in biological systems.
Michael Levin links.
Michael’s Twitter: https://twitter.com/drmichaellevin.
Michael’s Website: https://drmichaellevin.org.
PODCAST INFO:
The Learning With Lowell show is a series for the everyday mammal. In this show we’ll learn about leadership, science, and people building their change into the world. The goal is to dig deeply into people who most of us wouldn’t normally ever get to hear. The Host of the show – Lowell Thompson-is a lifelong autodidact, serial problem solver, and founder of startups.
LINKS
Youtube: https://www.youtube.com/channel/UCzri06unR-lMXbl6sqWP_-Q
Youtube clips: https://www.youtube.com/channel/UC-B5x371AzTGgK-_q3U_KfA
Linkedin: https://www.linkedin.com/in/lowell-thompson-2227b074
Twitter: https://twitter.com/LWThompson5
Website: https://www.learningwithlowell.com/
Shownotes/ Timestamps.
00:00 Introducing Michael Levin.
00:30 Epigenetic Head Exploding adaptation Planaria.
05:45 Generalize vs intelligent search epigenetic adaptation.
08:55 Designing studies to test these hypothesis.
12:35 Implications of hypothesis proven out.
19:40 Mitochondria domestication hypothesis.
25:50 Where are memories stored if not the brain.
34:45 Regeneration of memories evidence.
38:00 Voltage on both sides of amputated limb, and what catalyzes regeneration.
42:55 Induce physiology of extinct species from live species.
47:55 Biomanufacturing.
55:30 Anatomical compiler development.
57:45 Horse vs zebra domestication.
59:20 Bioelectricity resurrection.
01:02:05 Regeneration vs Brain computer interface for restoring function.
01:06:50 What is needed to achieve his vision for regeneration, bioelectricity, etc.
01:08:42 Structure needed to support development.
01:11:03 Groups coming together.
01:12:25 Longevity & health span — high level vs low level approach.
01:14:45 Cancer — why mortal cell become an immortal cell.
01:19:20 Advice for 25–35 year olds.
01:22:46 Age he discovered life goal.
01:23:55 How old he feels mentally.
01:24:55 Books.
01:25:55 Working to learn currently.
#Bioelectricity #MichaelLevin #Regeneration
A treatment that enables human beings to live to the age of 150 could be available within the next five to 10 years, one of the world’s most eminent plastic surgeons has claimed. Dr Steven Cohen, who specialises in complex aesthetic facial and regenerative surgery, believes the results of studies and advances in the medical sphere could soon pave the way to extend our lives.

Gene therapy has been headline news in recent years, in part due to the rapid development of biotechnology that enables doctors to administer such treatments. Broadly, gene therapies are techniques used to treat or prevent disease by tweaking the content or expression of cells’ DNA, often by replacing faulty genes with functional ones.
The term “gene therapy” sometimes appears alongside misinformation about mRNA vaccines, which include the Pfizer and Moderna COVID-19 vaccines. These vaccines contain mRNA, a genetic cousin of DNA, that prompts cells to make the coronavirus “spike protein.” The vaccines don’t alter cells’ DNA, and after making the spike, cells break down most of the mRNA. Other COVID-19 shots include the viral vector vaccines made by AstraZeneca and Johnson & Johnson, which deliver DNA into cells to make them build spike proteins. The cells that make spike proteins, using instructions from either mRNA or viral vector vaccines, serve as target practice for the immune system, so they don’t stick around long. That’s very, very different from gene therapy, which aims to change cells’ function for the long-term.
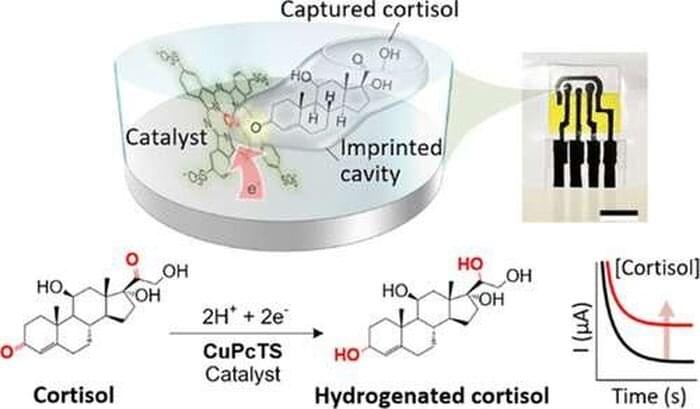
Researchers in the Oregon State University College of Engineering have developed a handheld sensor that tests perspiration for cortisol and provides results in eight minutes, a key advance in monitoring a hormone whose levels are a marker for many illnesses including various cancers.
Findings were published in the journal ACS Applied Materials & Interfaces. The material and sensing mechanism in the new device could be easily engineered to detect other specific hormones, the researchers say—for example, progesterone, a key marker for women’s reproductive health and pregnancy outcomes.
“We took inspiration from the natural enzymes used in blood glucose meters sold at pharmacies,” said Larry Cheng, associate professor of electrical engineering and computer science. “In glucose meters, specific enzymes are applied to an electrode, where they can capture and react with glucose molecules to generate an electrical signal for detection. However, finding natural enzymes for cortisol detection is not straightforward, and natural enzymes are prone to instability and have a short lifespan.”
Hailey-Hailey disease is a rare, inherited condition characterized by patches of blisters appearing mainly in the skin folds of the arm pits, groin and under the breasts. It is caused by a mutation in the gene that codes for a specific protein involved in the transportation of calcium and manganese ions from the cell cytoplasm and into a sac-like organelle called the Golgi apparatus.
Scientists at Tohoku University, together with colleagues in Japan, have uncovered some aspects of this protein’s structure that could help researchers understand how it works. The findings, published in the journal Science Advances, help build the foundations for research into finding treatments for Hailey-Hailey disease and other neurodegenerative conditions.
The protein the team studied is called secretory pathway Ca2+/Mn2+-ATPase, or SPCA for short. It is located in the Golgi apparatus, a cellular sac-like structure that plays a crucial role in protein quality control before they are released into cells. The Golgi apparatus also acts like a sort of calcium ion storage container. Calcium ions are vital for cell signaling processes and are important for proteins to function properly, so maintaining the right calcium ion balance inside cells is necessary for their day-to-day activities.
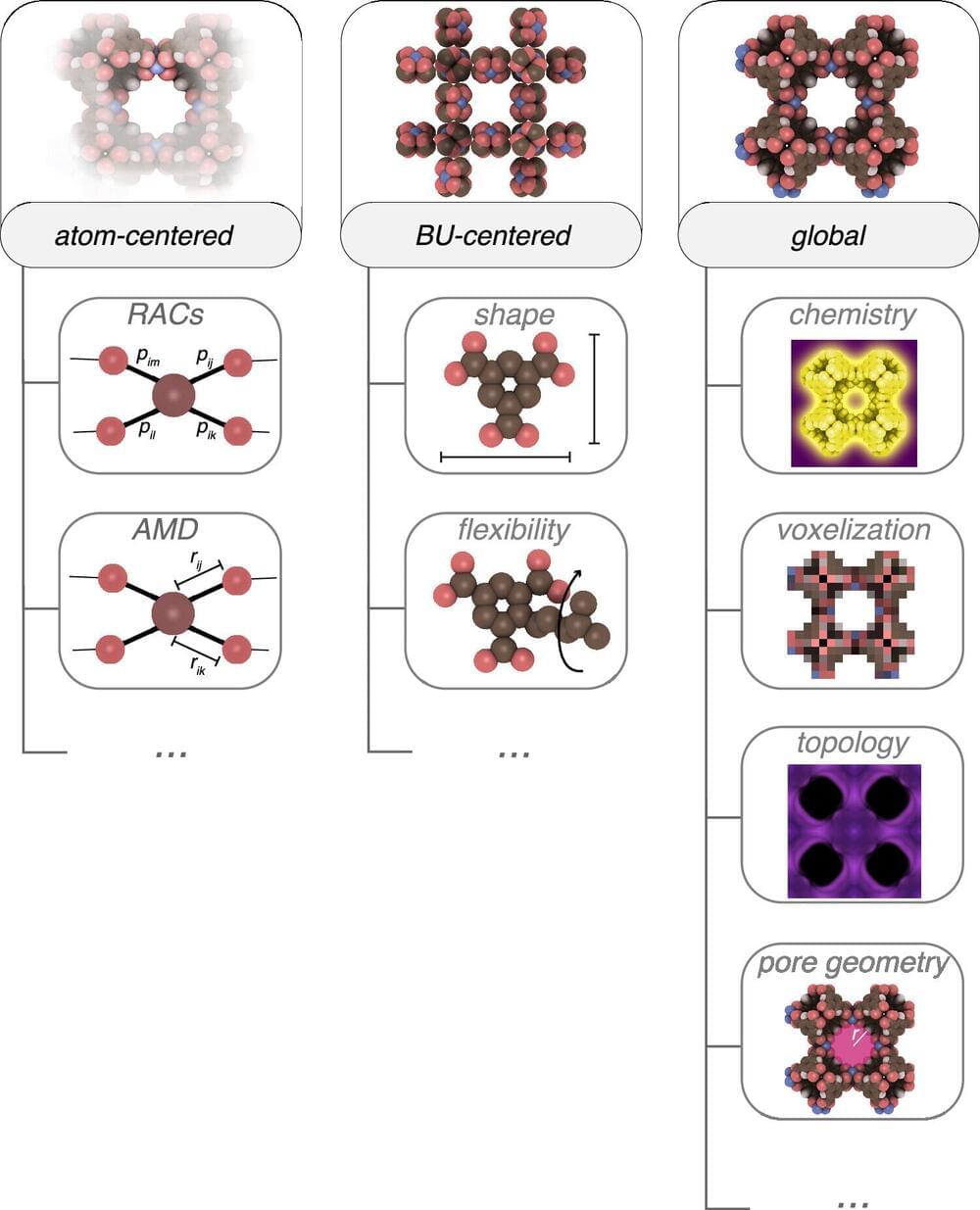
A team of chemists and computer scientists from the Swiss Federal Institute of Technology Lausanne, the University of California and Institut des Sciences et Ingenierie Chimiques, Ecole, have developed an ecosystem of tools to boost machine-learning-based design of metal-organic frameworks.
In their study, reported in the journal ACS Central Science, Kevin Maik Jablonka, Andrew Rosen, Aditi Krishnapriyan and Berend Smit coded tools to convert data into machine learning inputs to create a system to boost machine-learning frameworks.
Reticular chemistry is the science of designing and synthesizing porous crystalline materials with certain predefined structures and properties (building blocks). These materials, known as metal-organic frameworks (MOFs) have applications in gas storage, separation, catalysis, sensing and drug delivery.
Genetic Engineering extends far beyond the controversial news headlines that obsess over ‘designer babies’. In the science community, gene-editing tools like CRISPR and PRIME editing will do nothing less than save the planet.
The Rise Of Genetic Engineering (2022)
Writers: Kyle McCabe, Christopher Webb Young.
Stars: Rodolphe Barrangou, George Church, Mary Beth Dallas.
Genre: Documentary.
Country: United States.
Language: English.
Release Date: August 24, 2022 (United States)
Synopsis:
Genetic Engineering extends far beyond the controversial news headlines that obsess over ‘designer babies’. In the science community, gene-editing tools like CRISPR and PRIME editing will do nothing less than save the planet.
Methods like this allow scientists to alter and ‘re-program’ the genetics of living organisms.
This episode shows scientists at large using gene-editing technologies to revolutionize the food supply chain, bolstering food crops to prevent famines, and even speed up reforestation efforts that will reverse global warming. Genetic Engineering in farm animals is helping scientists to ‘select’ desirable traits, like physical features and gender. Incredibly, one scientist is using gene-editing technologies to resurrect the DNA of extinct species, like the Wooly Mammoth!
Despite some public concern, gene-editing is definitely a cause for hope in the fight against genetic disorders in humans. It’s already reversing a type of congenital blindness in children. And with the hyper-precision afforded by PRIME editing being prepared for clinical trials, a much more hopeful world will be revealed for families in the future.