Synthetic biology explained, future of synthetic biology, what is synthetic biology, creating life from DNA, synthetic life creation, synthetic biology techn…
Category: bioengineering – Page 28
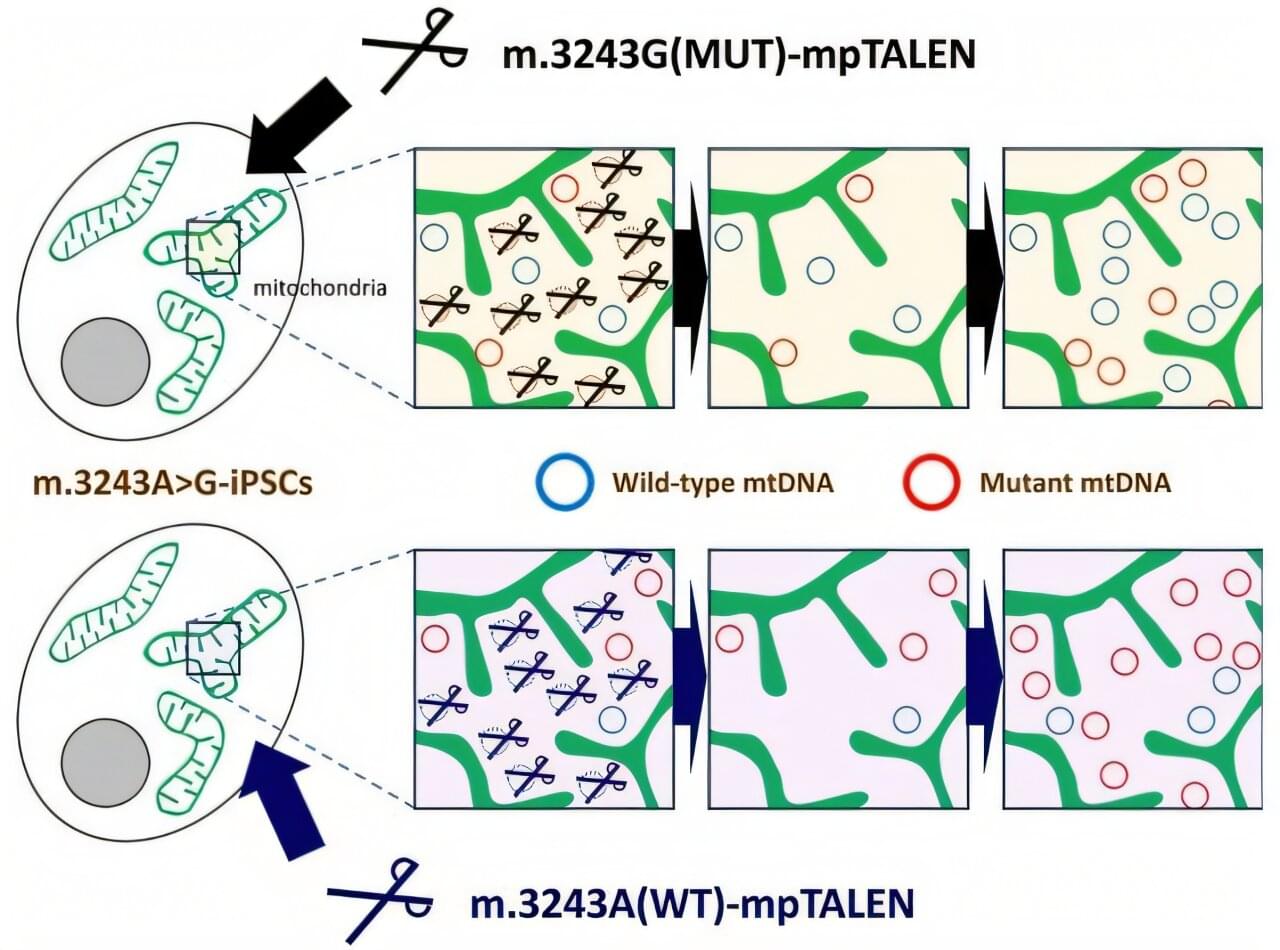
Engineered enzymes enable precise control of mitochondrial DNA mutation levels in cells
Mitochondrial diseases affect approximately 1 in 5,000 people worldwide, causing debilitating symptoms ranging from muscle weakness to stroke-like episodes. Some of these conditions result from mutations in mitochondrial DNA (mtDNA), the genetic material housed in these organelles. For patients with the common m.3243A>G mutation, which can cause MELAS syndrome (mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes) and diabetes mellitus, treatments remain limited.
Michael Levin: The New Era of Cognitive Biorobotics | Robinson’s Podcast #187
Patreon: https://bit.ly/3v8OhY7
Michael Levin is a Distinguished Professor in the Biology Department at Tufts University, where he holds the Vannevar Bush endowed Chair, and he is also associate faculty at the Wyss Institute at Harvard University. Michael and the Levin Lab work at the intersection of biology, artificial life, bioengineering, synthetic morphology, and cognitive science. Michael also appeared on the show in episode #151, which was all about synthetic life and collective intelligence. In this episode, Michael and Robinson discuss the nature of cognition, working with Daniel Dennett, how cognition can be realized by different structures and materials, how to define robots, a new class of robot called the Anthrobot, and whether or not we have moral obligations to biological robots.
The Levin Lab: https://drmichaellevin.org/
OUTLINE
00:00 Introduction.
02:14 What is Cognition?
08:01 On Working with Daniel Dennett.
13:17 Gatekeeping in Cognitive Science.
25:15 The Multi-Realizability of Cognition.
31:30 What are Anthrobots?
39:33 What Are Robots, Really?
59:53 Do We Have Moral Obligations to Biological Robots?
Robinson’s Website: http://robinsonerhardt.com
Robinson Erhardt researches symbolic logic and the foundations of mathematics at Stanford University. Join him in conversations with philosophers, scientists, weightlifters, artists, and everyone in-between.


Breakthrough DNA editing in Lactobacillus offers safer probiotics
A Kobe University team was able to edit the DNA of Lactobacillus strains directly without a template from other organisms. This technique is indistinguishable from natural variation and enabled the researchers to create a strain that doesn’t produce diabetes-aggravating chemicals.
Humans have improved the microorganisms we rely on for millennia, selecting variants that are better able to produce wine, yogurt, natto and many other products. More recently, direct genetic modification has emerged as a tool to exert more precise and efficient control over the improvement, but also has drawn much public criticism for often using DNA from unrelated organisms in these modifications. Kobe University bioengineer NISHIDA Keiji says, “As a consequence, using such transgenic techniques is not favorable for food products due to legislations being restrictive and social acceptance being low.”
Nishida and his team have developed a technique that gives even more precise control over the genetic content of a microorganism that does not rely on template DNA from other organisms. He says: “We have invented a DNA base editing technology named ‘Target-AID,’ which is superior to conventional techniques such as ‘CRISPR-Cas9’ in several aspects. For example, CRISPR-Cas9 induces DNA breaks and often causes cell death, while our Target-AID inserts precise point mutations without such breaks.”
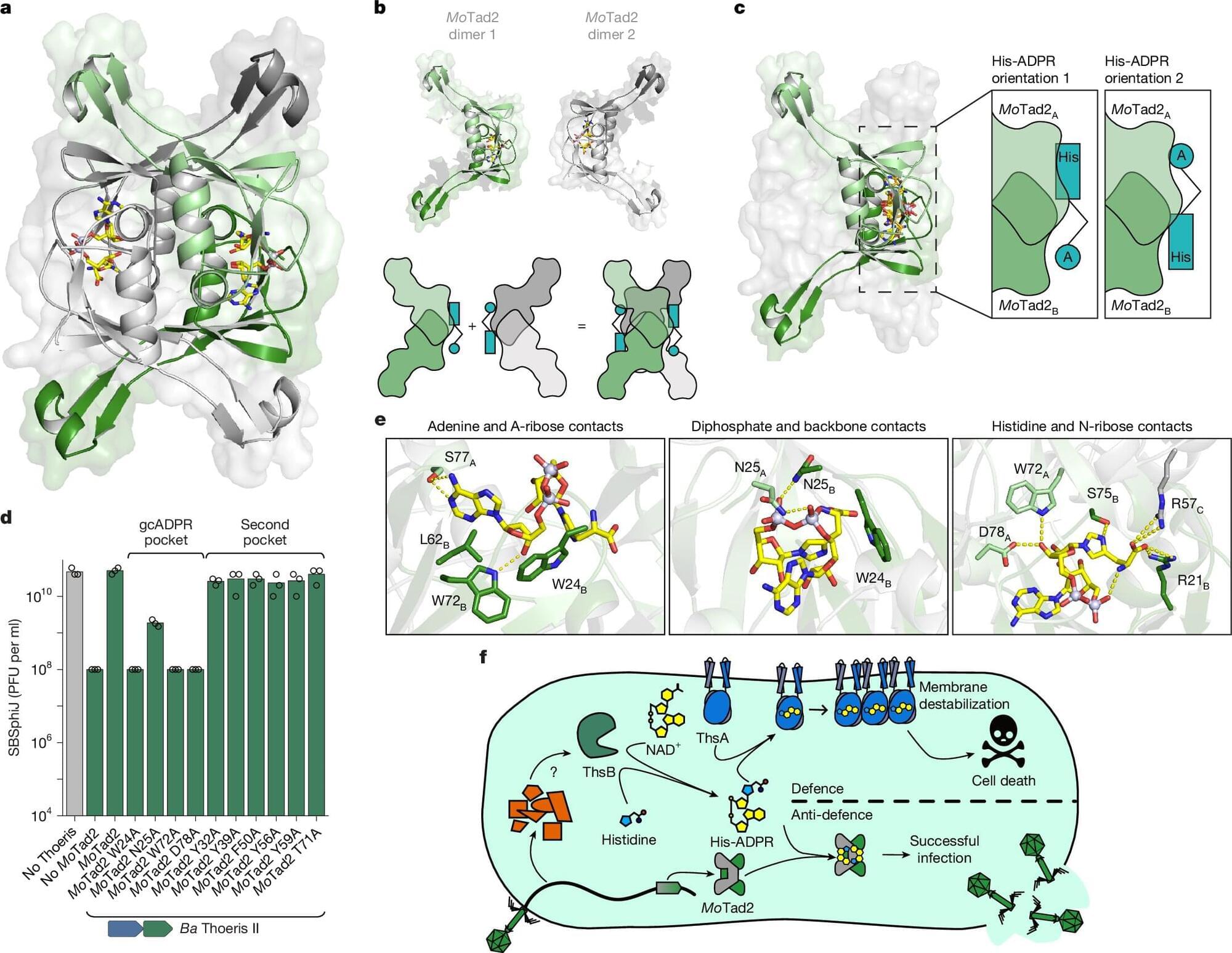
Discovery of histidine-ADP-ribose molecule reveals new bacterial immune strategy
A research team has uncovered a previously unknown type of immune signaling molecule—a novel compound combining histidine and ADP-ribose—produced by bacteria’s Thoeris II defense system in response to viral infection. This finding expands our understanding of bacterial immunity and may pave the way for innovative tools in biotechnology, gene editing, and antimicrobial therapy.
The paper, titled “TIR domains produce histidine-ADPR as an immune signal in bacteria,” is published in the journal Nature, and the team includes scientists at Vilnius University’s Life Sciences Centre (VU LSC), together with colleagues from the Weizmann Institute of Science (Israel) and Harvard Medical School.
The discovery sheds light on how bacteria, much like humans, communicate viral threats at the molecular level—in this case, triggering a self-sacrificing response to halt virus spread and protect bacterial populations. Beyond its fundamental significance, the finding opens exciting avenues for rethinking immune mechanisms and virus-host interactions.
Using Bacteria as Living Test Tubes to Study Human Gene Mutations and Find New Drug Leads
Traditional biochemical methods of studying human gene mutations are often laborious and costly. Now bioengineers at the University of California San Diego have developed a new simple approach to rapidly check on human gene changes and also screen chemicals as potential drugs by turning everyday bacteria into living test tubes.
The researchers published their new study in the April 30 issue of Nature Biomedical Engineering.
Human cells carry thousands of genes, and tiny changes in these genes can cause serious diseases. Usually, scientists study these changes by testing proteins in a test tube or in human cells. But those methods can be slow, expensive and sometimes hard to do.
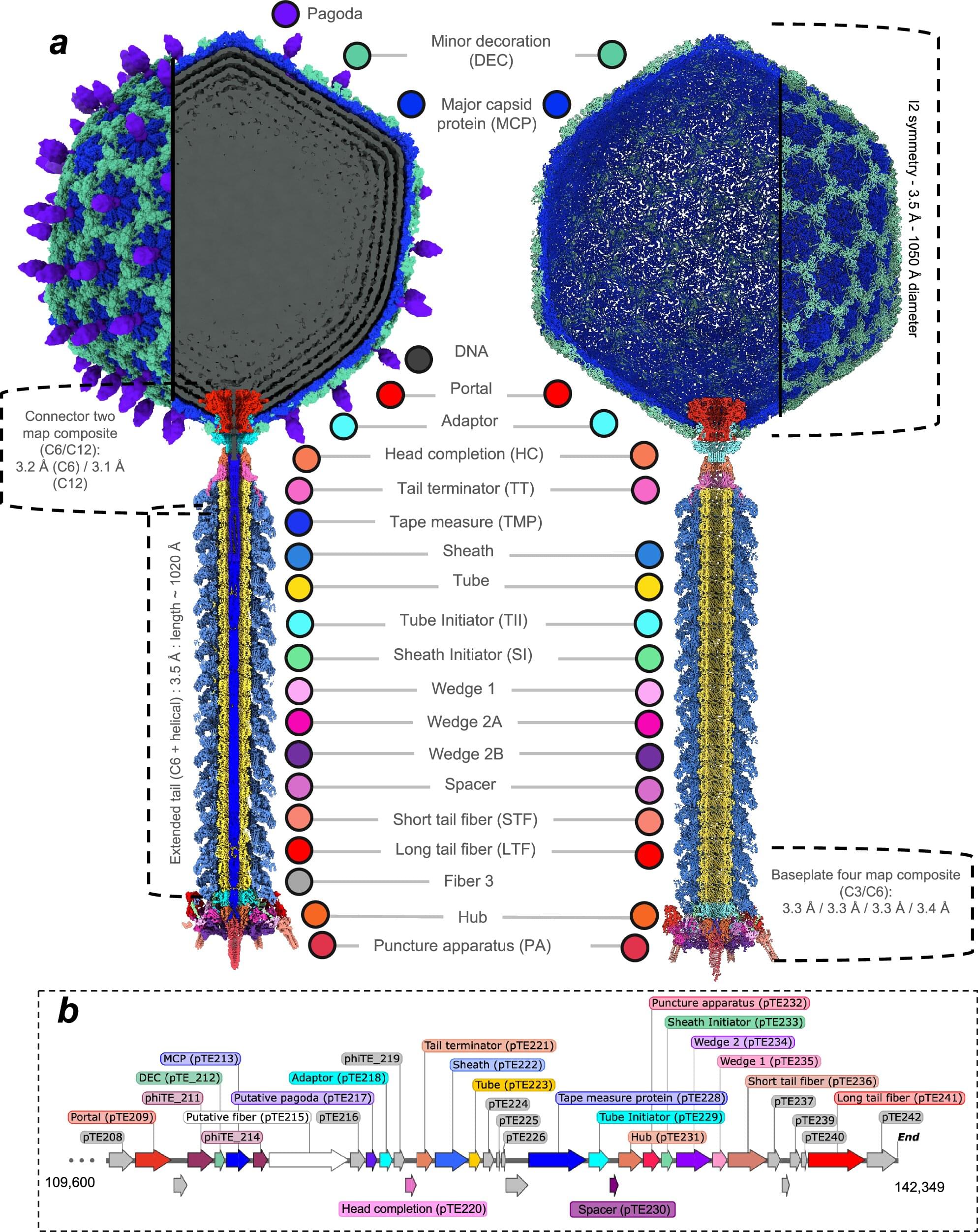
First atomic map of potato pathogen reveals potential infection mechanism
Plants are susceptible to a wide range of pathogens. For the common potato plant, one such threat is Pectobacterium atrosepticum, a bacterium that causes stems to blacken, tissues to decay, and often leads to plant death, resulting in significant agricultural losses each year.
In 2012, researchers isolated a new virus that infects and kills this bacterium—a bacteriophage named φTE (phiTE). Now, for the first time, scientists have uncovered the atomic structure of φTE, revealing a possible mechanism of infection that may be more complex than previously thought.
The study, published earlier this month in Nature Communications, is the result of a multidisciplinary collaboration between researchers from the Okinawa Institute of Science and Technology (OIST) and the University of Otago. It brings together expertise across several fields, including virology, structural biology, molecular genetics, protein engineering, biochemistry, and biophysics.
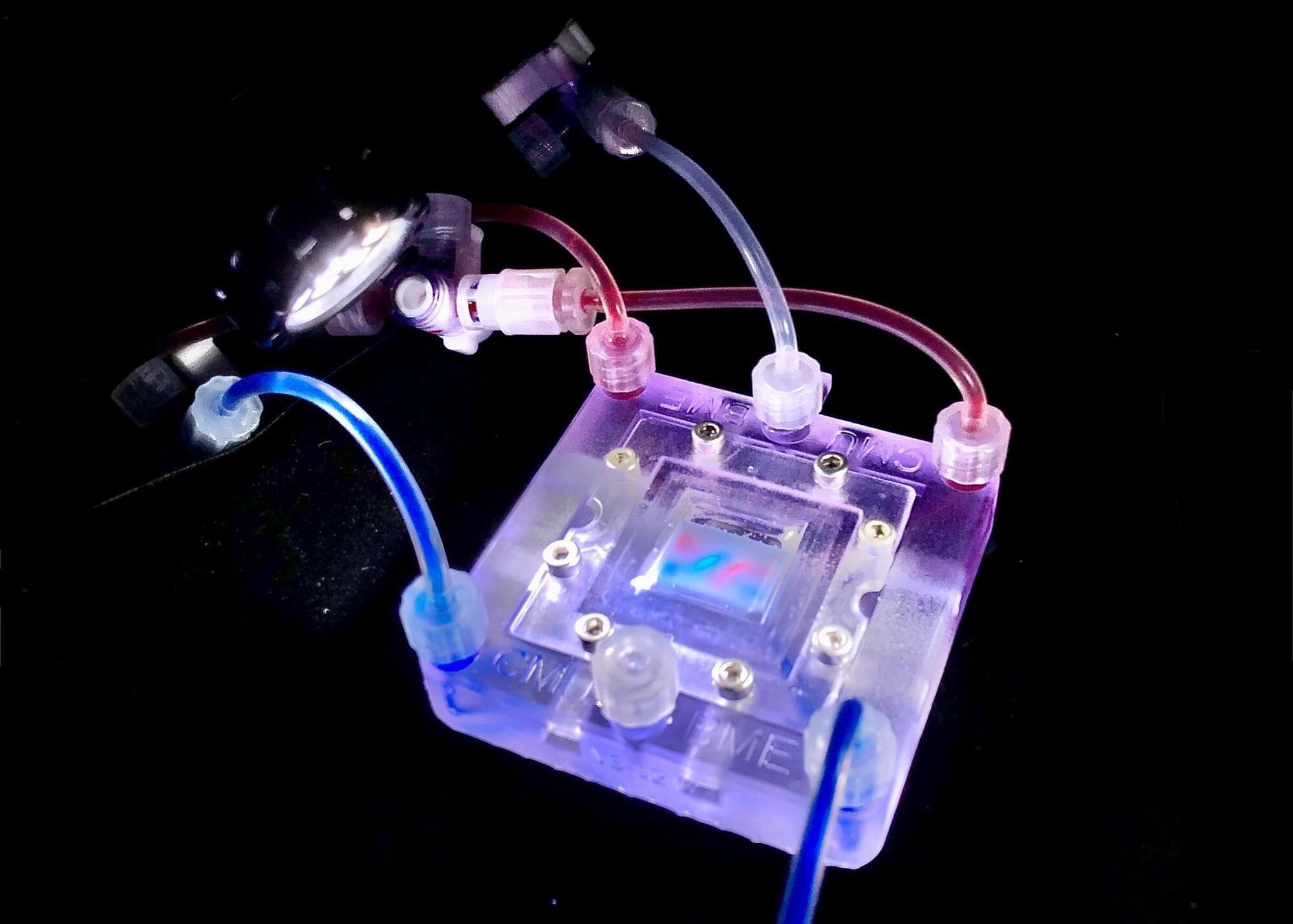
Printing the Future of Life: How 3D Collagen Scaffolds Grow Real Tissues
Researchers at the University of Pittsburgh have created a groundbreaking tissue engineering platform using 3D-printed collagen scaffolds called CHIPS.
By mimicking natural cellular environments, they enable cells to grow, interact, and form functional tissues — a major step beyond traditional silicone-based microfluidic models. The platform not only models diseases like diabetes but could also replace animal testing in the future. Plus, their designs are freely available to fuel broader scientific innovation.
3D bioprinting: turning science fiction into science reality.
