
Get the latest international news and world events from around the world.


NASA’s Hubble Space Telescope
It’s estimated that about 100 million black holes roam around our Milky Way Galaxy — and for the first time ever, astronomers now believe they may have precisely measured the mass of an isolated black hole with Hubble.
Roaming black holes are born from rare, monstrous stars that are at least 20 times more massive than our Sun. After these stars explode in a supernova, the remnant core is crushed by gravity into a black hole. Because this self-detonation isn’t perfectly symmetrical, the black hole might get “kicked” and careen through our galaxy.
Astronomers believe that the isolated black hole measured by Hubble is traveling across the Milky Way at 100,000 miles per hour (160,000 kph). That’s fast enough to get from Earth to the Moon in less than three hours!
Read more about this discovery: https://go.nasa.gov/3mx6t6p.
#NASA #Hubble #BlackHole #astronomy #science #universe #astrophysics #space #stars #galaxy
View insights.


Meet the Giant Sequoia, the ‘Super Tree’ Built to Withstand Fire
Mammoth redwood trees have evolved along with fire, but humans are disrupting that delicate balance.


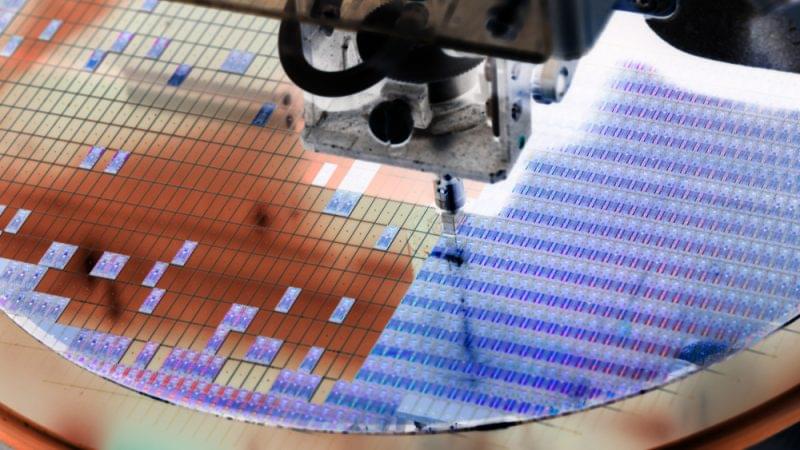
France welcomes new chips production site in bid to become global player
France is set to welcome a new semiconductor production site on its soil as it continues the drive to position itself on the global market amid growing shortages, though it is still unclear where funding for the new plant is coming from.
Read the original French article here.
France will open a new semiconductor factory, according to the announcement by French company STMicroelectronics and US company GlobalFoundries released on Monday (11 July).

Ten years after the Higgs, physicists face the nightmare of finding nothing else
Unless Europe’s Large Hadron Collider coughs up a surprise, the field of particle physics may wheeze to its end.
CELESTA, the first CERN-driven satellite, successfully entered orbit during the maiden flight of Europe’s Vega-C launch vehicle. Launched by the European Space Agency from the French Guiana Space Centre (CSG) at 13.13 UTC on 13 July 2022, the satellite deployed smoothly and transmitted its first signals in the afternoon. Weighing one kilogram and measuring 10 centimetres on each of its sides, CELESTA (CERN latchup and radmon experiment student satellite) is a 1U CubeSat designed to study the effects of cosmic radiation on electronics. The satellite carries a Space RadMon, a miniature version of a well-proven radiation monitoring device deployed in CERN’s Large Hadron Collider (LHC). CELESTA has been sent into an Earth orbit of almost 6,000 kilometres.