The experiment is a collaboration between the University of Cambridge and Université Libre de Bruxelles – partner institutions of the European Commission’s Graphene Flagship – along with the Mohammed bin Rashid Space Centre (MBRSC) in the United Arab Emirates, York University in Canada, and the European Space Agency (ESA).
Regolith is composed of extremely sharp, tiny and sticky grains and, since the Apollo missions, it has been one of the biggest challenges lunar missions have had to overcome. Regolith can cause mechanical and electrostatic damage to equipment and is therefore also hazardous for astronauts. It clogs spacesuits’ joints, obscures visors, erodes spacesuits and protective layers, and is a potential health hazard.
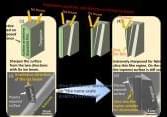
Cambridge researchers have produced special graphene composites that are meant to reduce regolith adhesion. The graphene samples will be monitored via an optical camera, which will record footage throughout the mission. Researchers from Université Libre de Bruxelles (ULB) will gather information during the mission and suggest adjustments to the path and orientation of the rover. Data and images obtained will be used to study the effects of the moon environment and the regolith abrasive stresses on the samples.