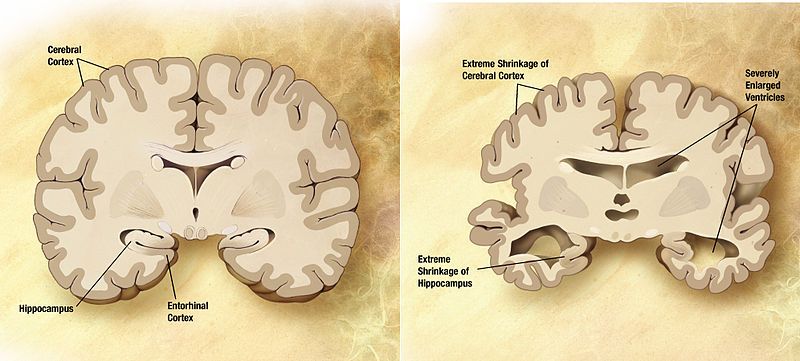
The core of what we do at Nanalyze is to tell our readers all they need to know about investing in emerging technologies. Sometimes that story is much, much bigger, and what we’re really talking about is investing in emerging industries. NewSpace is one example, launching about 15 years ago with the emergence of companies like SpaceX and Virgin Galactic. It’s probably only within the last five years that the NewSpace industry has achieved real liftoff, with dozens of startups doing everything from offering launch services to building satellites to developing business analytics from space-based imagery. While we may one day end up living on Mars, we’re more interested in living a long and fruitful life right here on Mother Earth, despite the specter of cancer and dementia. An entire industry is coalescing around human longevity, promising to beat these age-related diseases and extend our lives to biblical proportions.
We’ve been covering the topic of life extension for more than five years, beginning with a profile on an anti-aging company called Human Longevity Inc, whose founders include billionaire serial entrepreneur Peter Diamandis and J. Craig Venter, a leading genomics expert. More recently, we introduced you to nine companies developing products in regenerative medicine, a broad category that refers to restoring the structure and function of damaged tissues or organs. We also tackled the more controversial topic of young blood transfusions earlier this year, as well as covered the 2019 IPO of Precision BioSciences (DTIL), a gene-editing company that wants to fight disease and re-engineer food.