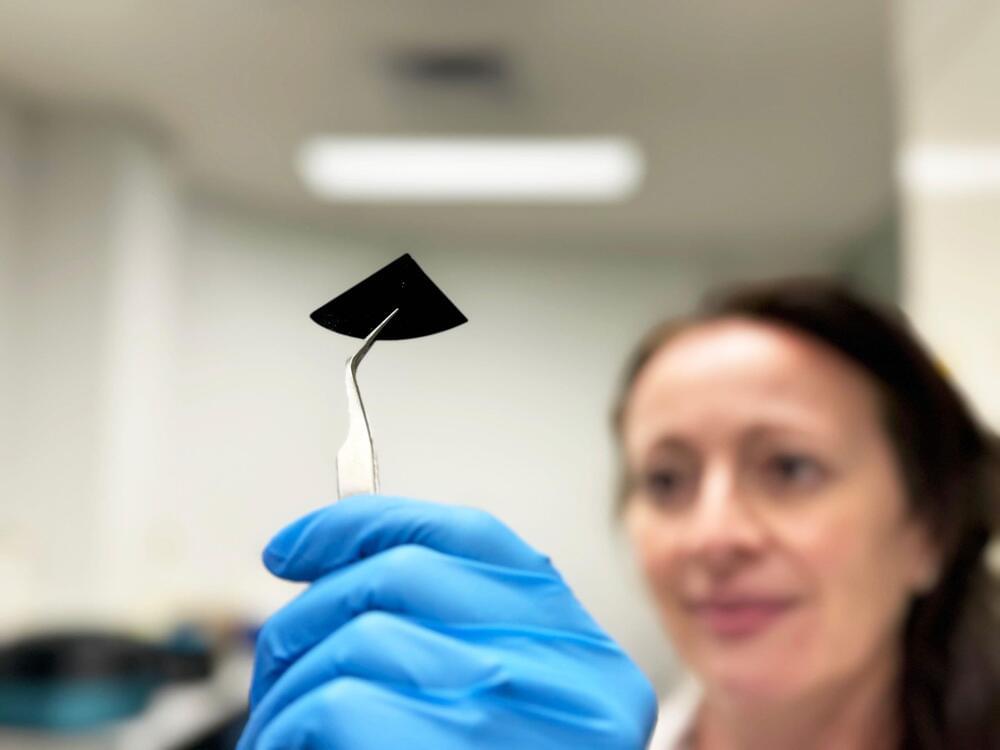
Leafhoppers, a common backyard insect, secrete and coat themselves in tiny mysterious particles that could provide both the inspiration and the instructions for next-generation technology, according to a new study led by Penn State researchers. In a first, the team precisely replicated the complex geometry of these particles, called brochosomes, and elucidated a better understanding of how they absorb both visible and ultraviolet light.
This could allow the development of bioinspired optical materials with possible applications ranging from invisible cloaking devices to coatings to more efficiently harvest solar energy, said Tak-Sing Wong, professor of mechanical engineering and biomedical engineering. Wong led the study, which was published today (March 18) in the Proceedings of the National Academy of Sciences of the United States of America (PNAS).