While the artificial intelligence revolution has just begun, it is transforming healthcare, speeding drug discovery, improving both diagnosis and patient communication.


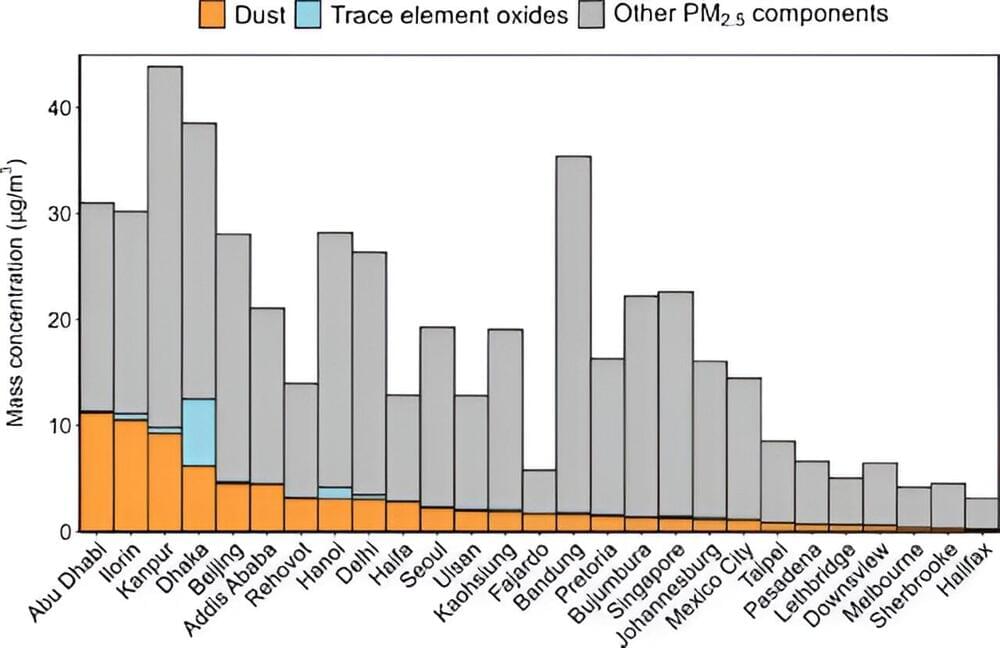
As anyone with seasonal allergies knows, unseen airborne particles can really wreck a person’s day. Like the tree pollen that might be plaguing you this spring, small concentrations of trace elements in the air can have significant negative impacts on human health. However, unlike pollen counts and other allergy indices, which are carefully tracked and widely available, limited knowledge exists about the ambient concentrations of cancer-causing trace elements like lead and arsenic in urban areas of developing countries.
An innovative programmable tool for targeting nucleic acids has been created, utilizing a prokaryotic immune defense system—and it is not CRISPR-Cas. Russian Academy of Sciences researchers have successfully re-engineered prokaryotic Argonautes (pAgos) to utilize RNA guides for locating nucleic acid sequences. These systems have been modified to form a complex with effector nucleases.
The researchers employed a two-component system known as SPARDA (short prokaryotic Argonaute, DNase, and RNase-associated) to effectively identify DNA sequences with a notable level of sensitivity and induce collateral nuclease activity. SPARDA and other concise pAgos systems that encode diverse effectors have the potential to offer a novel programmable tool for the field of biotechnology.
The research article “DNA-targeting short Argonautes complex with effector proteins for collateral nuclease activity and bacterial population immunity” was published in Nature Microbiology.
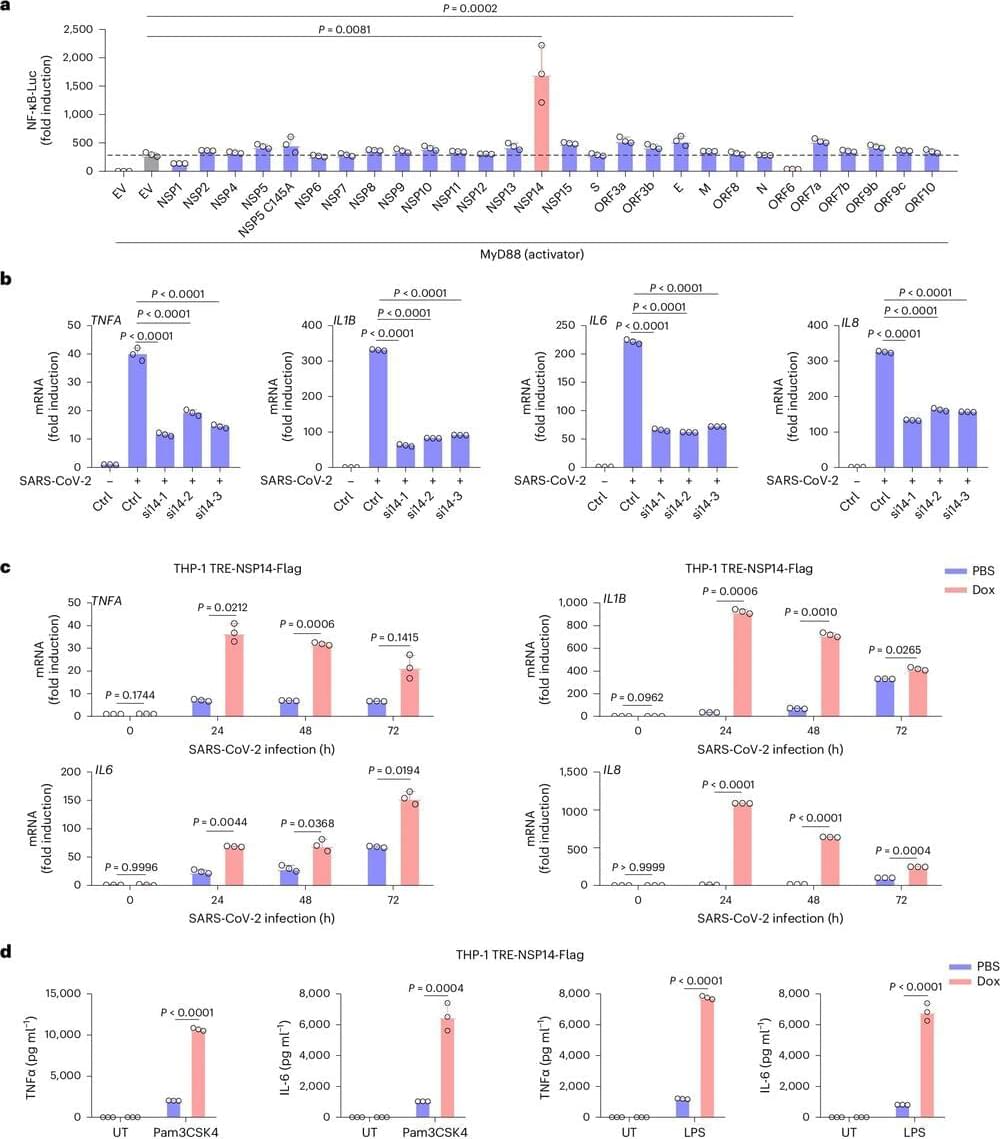
A recent USC study provides new information about why SARS-CoV-2, the virus behind the COVID-19 pandemic, may elicit mild symptoms at first but then, for a subset of patients, turn potentially fatal a week or so after infection. The researchers showed that distinct stages of illness correspond with the coronavirus acting differently in two different populations of cells.
The study, published in Nature Cell Biology, may provide a roadmap for addressing cytokine storms and other excessive immune reactions that drive serious COVID-19.
The team found that when SARS-CoV-2 infects its first-phase targets, cells in the lining of the lung, two viral proteins circulate within those cells—one that works to activate the immune system and a second that, paradoxically, blocks that signal, resulting in little or no inflammation.
I found this on NewsBreak.
According to reports, Japanese scientists have devised a technique for connecting lab-grown brain-mimicking tissue„ like how circuits in our brain work.
Researchers at the University of Tokyo released a study in Nature Communications journal that looked into making a seemingly impossible idea a reality.
The scientists discovered a new approach to establishing physiological connections between lab-grown neuronal organoids. These organoids are experimental model tissues created by growing human stem cells into 3D-developed brain-like structures.

Zocdoc: Go to https://www.zocdoc.com/ICED and download the Zocdoc App for FREE NetSuite: Take advantage of NetSuite’s FREE KPI checklist: https://www.netsuite.com/ICED Insider Clothing: Head to https://insider.clothing/IcedCoffeeHour and use code ICED15 for 15% off your order Hims: Start your free online visit today at https://hims.com/ich Follow Dr. Alok Kanojia and subscribe to him here: https://www.youtube.com/@HealthyGamerGG Want to understand your mental health? Check out Dr. K’s Guides to Mental Health https://bit.ly/4caPuiC Dr. K’s Book, How to Raise a Healthy Gamer https://bit.ly/3Pk8D8e NEW: Join us at http://www.icedcoffeehour.club for premium content — Enjoy! Add us on Instagram: https://www.instagram.com/jlsselby https://www.instagram.com/gpstephan Official Clips Channel: https://www.youtube.com/channel/UCeBQ24VfikOriqSdKtomh0w For sponsorships or business inquiries reach out to: [email protected] For Podcast Inquiries, please DM @icedcoffeehour on Instagram! Time Stamps: 0:00 — Intro 0:52 — This Is The RIGHT Way To Sit 6:59 — How To Know You’re ACTUALLY Happy 19:34 — How 99% of Things Are Out Of Your Control 24:13 — Who Is Dr. K? (Background) 26:18 — There Is No Such Thing As Good OR Bad 31:18 — Should You Go To Therapy? 33:26 — Dr. K’s Thoughts On Tony Robbins & Neuro-Linguistic Programming 39:03 — How To ACTUALLY Become Happy 1:02:33 — How Much Sacrifice Is Required To Be Successful? 1:14:09 — How To Get Into Your Flow State At Work 1:22:25 — Why Dr. K Thinks ‘Monk Mode’ is “Silly” 1:31:07 — Dr. K Explains Burn Out 1:39:07 — How Our Brains Can Experience “Hypothetical Pain” As REAL PAIN 1:42:47 — How To See NEGATIVES As POSITIVES 2:00:46 — Dr. K Explains The Whole Scale FAILURE of Our Traditional Institutions 2:16:26 — How Our Minds Are Being Controlled 2:46:28 — Is Social Media A GOOD or BAD Thing Overall? 2:49:24 — Dr. K’s Thoughts On Drama Bait YouTube Channels & Instagram ‘Gore” Reels 2:57:27 — Dr. K On PORN & INCELS 3:13:19 — Should You Make Decisions For Your Significant Other? 3:17:41 — Why People Lie & The POWER Of Truth *Emotional* 3:28:30 — How Terminal Patients Learn How To Deal W/ Death 3:35:34 — How Dr. K Personally Deals With Trauma & Negativity 3:59:07 — Dr. K Brings Jack & Graham Through A Meditation Exercise 4:15:31 — Closing Thoughts *Some of the links and other products that appear on this video are from companies which Graham Stephan will earn an affiliate commission or referral bonus.
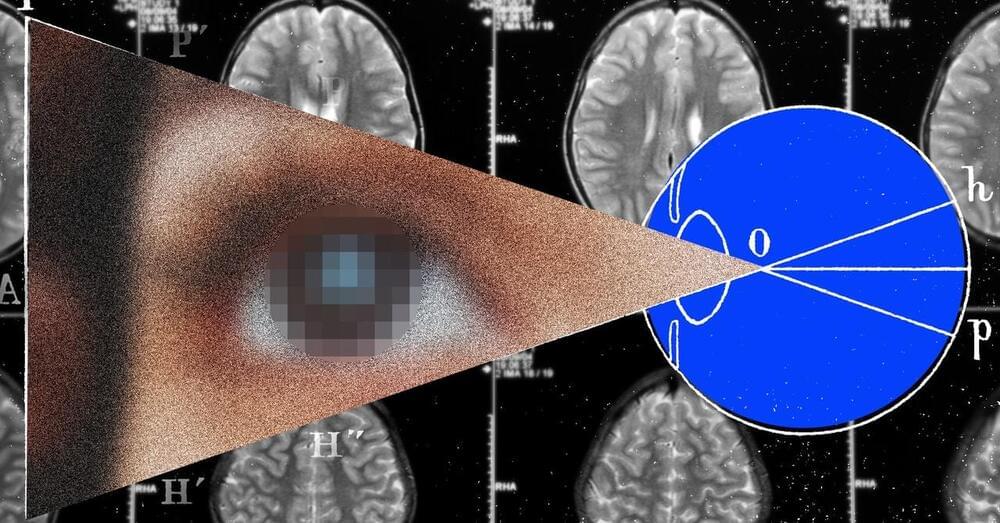
In 2021, he heard about a trial of a visual prosthesis at Illinois Institute of Technology in Chicago. Researchers cautioned that the device was experimental and he shouldn’t expect to regain the level of vision he had before. Still, he was intrigued enough to sign up. Thanks to the chips in his brain, Bussard now has very limited artificial vision—what he describes as “blips on a radar screen.” With the implant, he can perceive people and objects represented in white and iridescent dots.
Bussard is one of a small number of blind individuals around the world who have risked brain surgery to get a visual prosthesis. In Spain, researchers at Miguel Hernández University have implanted four people with a similar system. The trials are the culmination of decades of research.
There’s interest from industry, too. California-based Cortigent is developing the Orion, which has been implanted in six volunteers. Elon Musk’s Neuralink is also working on a brain implant for vision. In an X post in March, Musk said Neuralink’s device, called Blindsight, is “already working in monkeys.” He added: “Resolution will be low at first, like early Nintendo graphics, but ultimately may exceed normal human vision.”
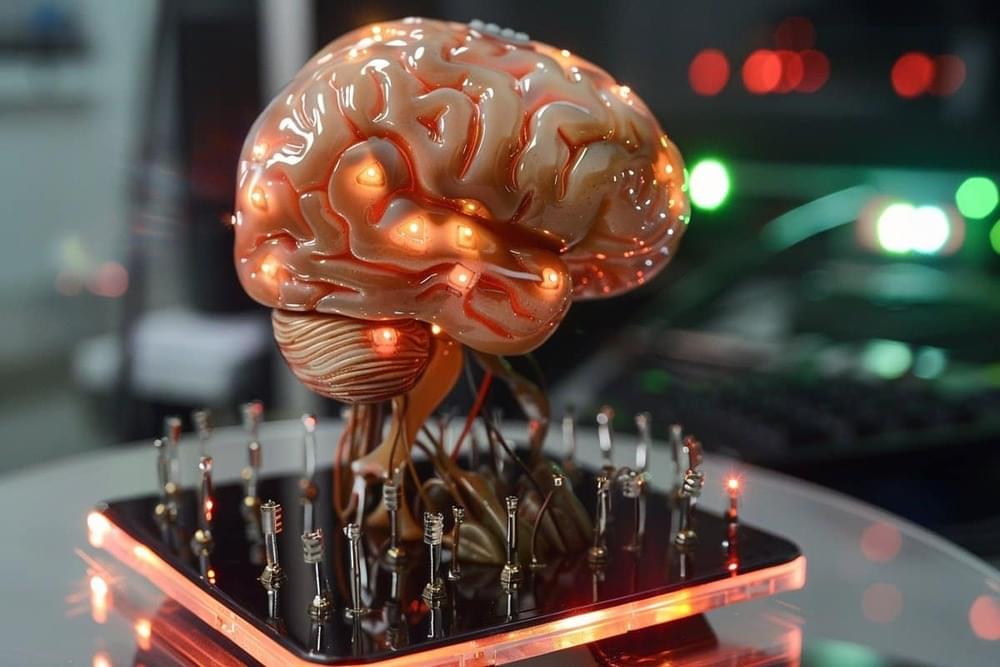
Summary: Researchers developed a groundbreaking pea-sized brain stimulator, the Digitally Programmable Over-brain Therapeutic (DOT), capable of wireless operation through magnetoelectric power transfer. This implantable device promises to revolutionize treatment for neurological and psychiatric disorders by enabling less invasive and more autonomous therapeutic options compared to traditional neurostimulation methods.
The DOT’s ability to stimulate the brain through the dura without implanted batteries represents a significant advancement in medical technology, offering potential treatments for conditions like drug-resistant depression directly from the comfort of one’s home. This innovation could change the landscape of how brain-related disorders are managed, emphasizing patient comfort and control.