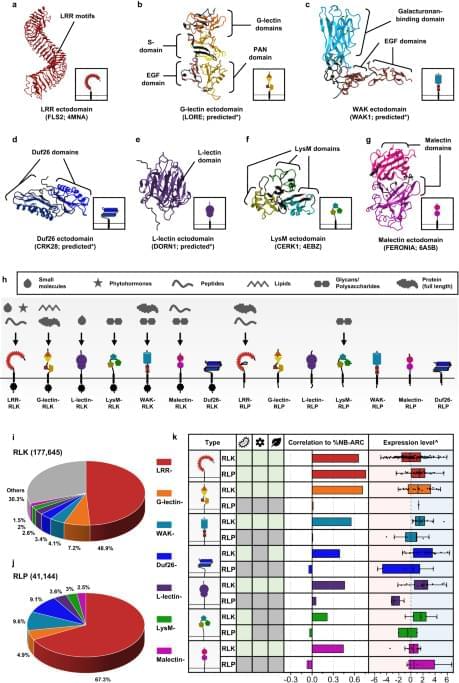
Plant cell-surface receptors that are known to participate in immunity, development, and reproductive processes include the LRR-, G-lectin-, Wall-associated kinase (WAK)-, Domain of Unknown Function 26 (Duf26)-, L-lectin-, Lysin motif (LysM)-, and Malectin-containing RLKs and RLPs (Fig. 1a–h). There are additional RLK families with different ectodomains, such as the proline-rich extensin-like receptor kinases (PERKs) and thaumatin-like protein kinases (TLPKs)9,13. However, their function in immunity is not well-characterized. Cell-surface receptors with LRR-, G-lectin-, WAK-, and LysM-ectodomains have been reported to recognise PAMPs, while others perceive self-molecules or unidentified ligands (Fig. 1h; Supplementary Fig. 1). Recognition of the diverse array of ligands is likely to be accomplished by variable structures and combinations of different ectodomains (Fig. 1a–g). To trace the origins of different receptor classes within the plant lineage, we first identified RLKs and RLPs in 350 genomes from Glaucophyta, red algae, green algae, Bryophytes, and Tracheophytes. We define here RLKs as any proteins with both 1–2 TMs and KDs, and RLPs as any protein with 1–2 TMs, but lack KDs. In total, we identified 177,645 RLKs, almost up to 70% of which possess either LRR-, G-lectin-, WAK-, Duf26-, L-lectin-, LysM-and Malectin-ectodomains (Fig. 1i). Next, we searched for proteins with these ectodomains and TMs that lack KDs and found 41,144 RLPs (Fig. 1j). We further examined which of the identified RLKs and RLPs families are likely to be involved in immunity. A previous report suggested a positive correlation between the gene family sizes of cell-surface immune receptors and intracellular immune receptors (the NB-ARC family) across the angiosperms4. We examined the correlation between the relative size (%; number of identified genes in the family/numbers of searched genes × 100; see methods) of the RLK families, the RLPs families, and the NB-ARC family in each genome. Notably, most RLK families (except for the LysM-RLKs) exhibit positive correlations with the NB-ARC family, while most RLP families (except for the LRR-RLPs) do not exhibit positive correlation with the NB-ARC family (Main Fig. 1k). Furthermore, we checked the expression level of these receptor families in Arabidopsis thaliana during immunity. Notably, the RLKs, except for LRR-and Malectin-RLKs, generally exhibit higher expression levels compared to the RLPs during immunity (Main Fig. 1k; Supplementary Fig. 2). These data collectively suggest that the RLKs are more likely to be involved in immunity than the RLPs.
Next, we examined the presence or absence of ectodomains (LRR-, G-lectin-, WAK-, Duf26-, L-lectin-, LysM-and Malectin-ectodomains lacking TM or KD; ectodomain-only proteins), RLPs (TM-bound ectodomains) and RLKs (ectodomains encompassing both TM and KD) in the plant lineage (Fig. 2; Supplementary Fig. 3; Supplementary Data 1a–c). Ectodomains exhibit an ancient heritage, with LRR-, WAK-, LysM-, Malectin-, and L-lectin-domains dating back to the era of Glaucophyta. Similarly, relatively ancient counterparts such as LRR-RLPs, WAK-RLPs, LysM-RLPs, Malectin-RLPs, and L-lectin-RLPs are found in both Glaucophyta and Rhodophyta. In contrast, RLKs emerged more recently. Green algae harbour WAK-RLKs, Malectin-RLKs, and G-lectin-RLKs, and LysM-RLKs, L-lectin-RLKs, and Duf-26-RLKs are exclusive to Embryophytes (Fig. 2). Except for LRR-RLPs, all six families of RLP are basal to the RLK families.