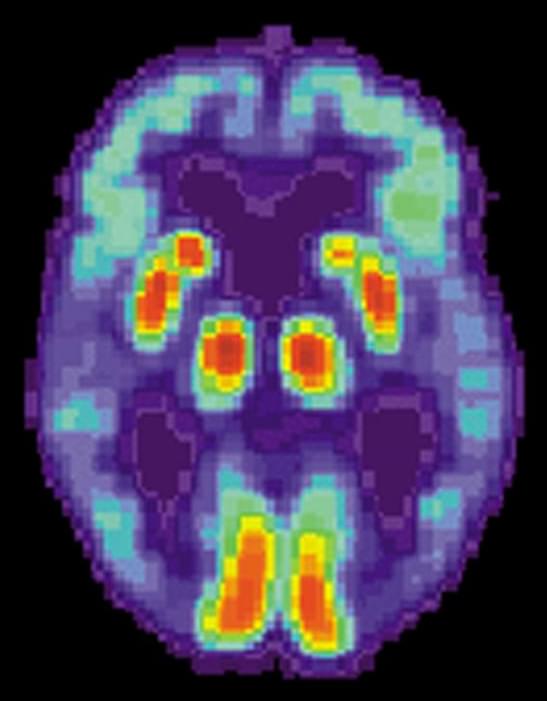
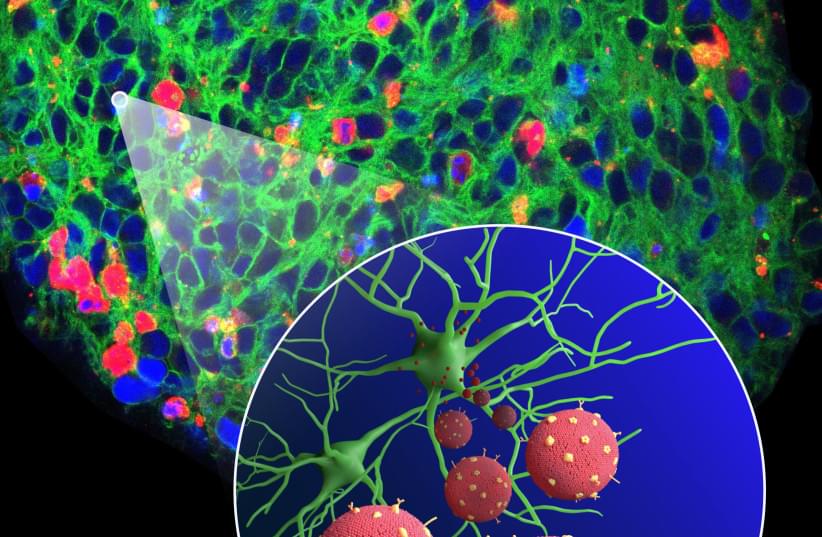
Two drugs approved decades ago not only counteract brain damage caused by Alzheimer’s disease in animal models, the same therapeutic combination may also improve cognition.
Sounds like a slam dunk in terms of a cure—but not yet. Researchers currently are concentrating on animal studies amid implications that remain explosive: If a surprising drug combination continues to destroy a key feature of the disease, then an effective treatment for Alzheimer’s may have been hiding for decades in plain sight.
A promising series of early studies is highlighting two well known medicine cabinet standbys—gemfibrosil, an old-school cholesterol-lowering drug, and retinoic acid, a vitamin A derivative. Gemfibrosil, is sold as Lopid and while it’s still used, it is not widely prescribed. Doctors now prefer to prescribe statins to lower cholesterol. Retinoic acid has been used in various formulations to treat everything from acne to psoriasis to cancer.