A laboratory study reveals an interaction between dietary fiber and the gut microbiome that may be helpful for fighting cancer.

Scientists may have uncovered a surprising new weapon in the fight against diabetes: the fruit of an ancient desert plant. Known as Nitraria roborowskii Kom, this resilient shrub has long been used in traditional medicine but has only recently gained scientific attention. In modern experiments, its fruit extract displayed a remarkable ability to reverse insulin resistance and restore healthy metabolism in diabetic mice.
The results went far beyond stabilizing blood sugar. Researchers found that the extract also corrected lipid imbalances and reduced oxidative stress, two major complications of diabetes. These effects were linked to the activation of a key cellular signaling pathway that helps regulate metabolism. Taken together, the findings suggest a powerful new direction for developing safer, more natural treatments for diabetes, one of the world’s most common chronic diseases.
As the number of people living with diabetes continues to climb, potentially reaching 750 million by 2045, scientists are urgently seeking better ways to manage the condition. Current medications can treat symptoms but often fail to tackle the root causes of metabolic dysfunction and may cause side effects over time. This challenge has driven researchers to reexamine traditional medicines in search of untapped therapeutic compounds.



AI has emerged as a pivotal tool in redefining TBI rehabilitation, bridging gaps in traditional care with innovative, data-driven approaches. While its potential to enhance diagnostic accuracy, outcome prediction, and individualized therapy is evident, challenges such as bias in datasets and ethical implications must be addressed. Continued research and multidisciplinary collaboration will be key to harnessing AI’s full potential, ensuring equitable access and optimizing recovery outcomes for TBI patients.
Overall, the integration of AI in TBI rehabilitation presents numerous opportunities to advance patient care and enhance the effectiveness of therapeutic interventions.
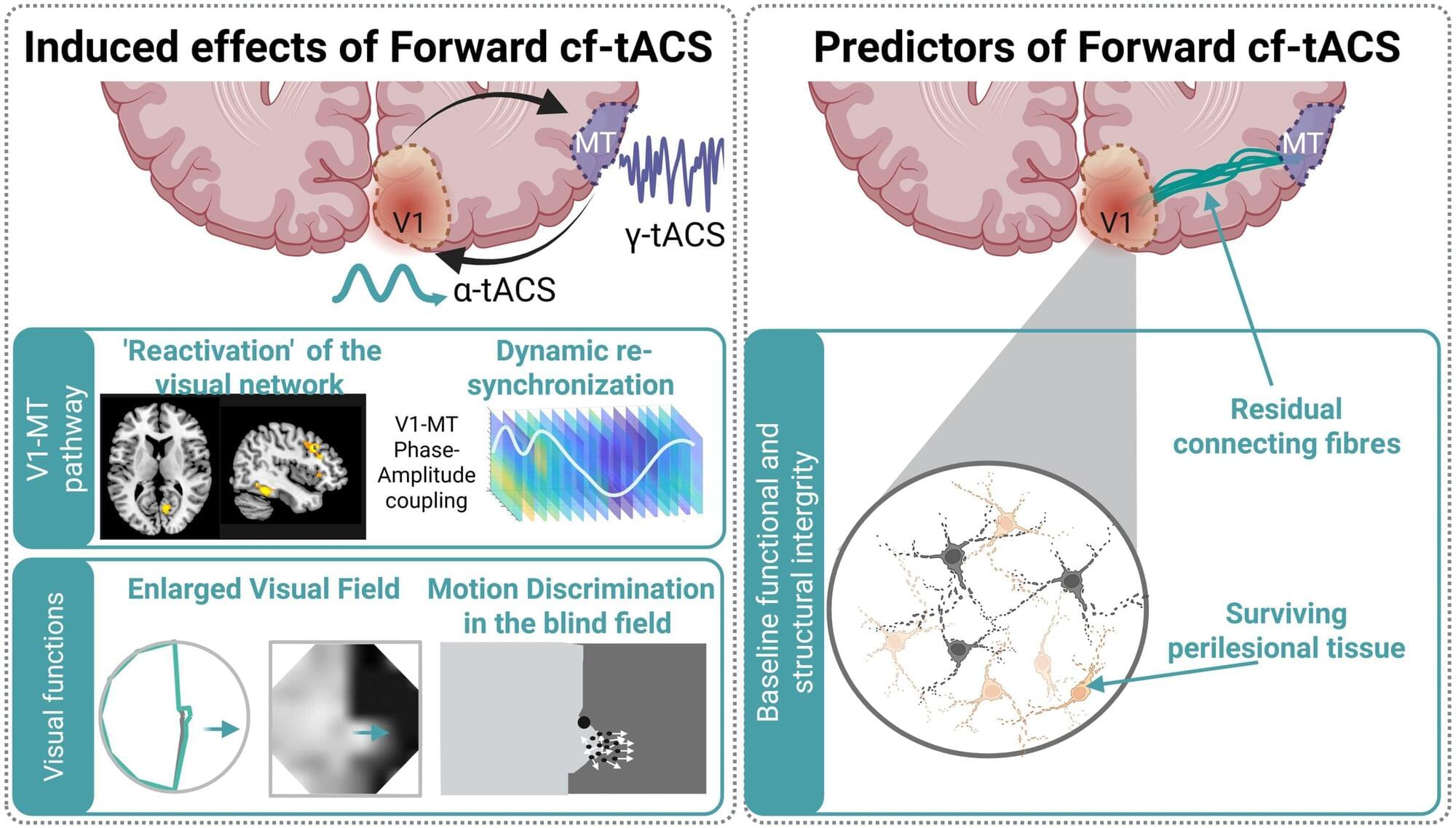
Scientists at EPFL have developed an innovative, non-invasive brain stimulation therapy to significantly improve visual function in stroke patients who have suffered vision loss following a stroke. The approach could offer a more efficient and faster way to regain visual function in such cases.
Each year, thousands of stroke survivors are left with hemianopia, a condition that causes loss of half of their visual field (the “vertical midline”). Hemianopia severely affects daily activities such as reading, driving, or just walking through a crowded space.
There are currently no treatments that can restore lost visual function in hemianopia satisfactorily. Most available options focus on teaching patients how to adapt to loss of vision rather than recovering it. To achieve some degree of recovery, months of intensive neurorehabilitative training are required for only moderate restoration at best.
This issue’s cover features companion papers that exemplify how understanding a rare disease can inform treatment strategies in other conditions.
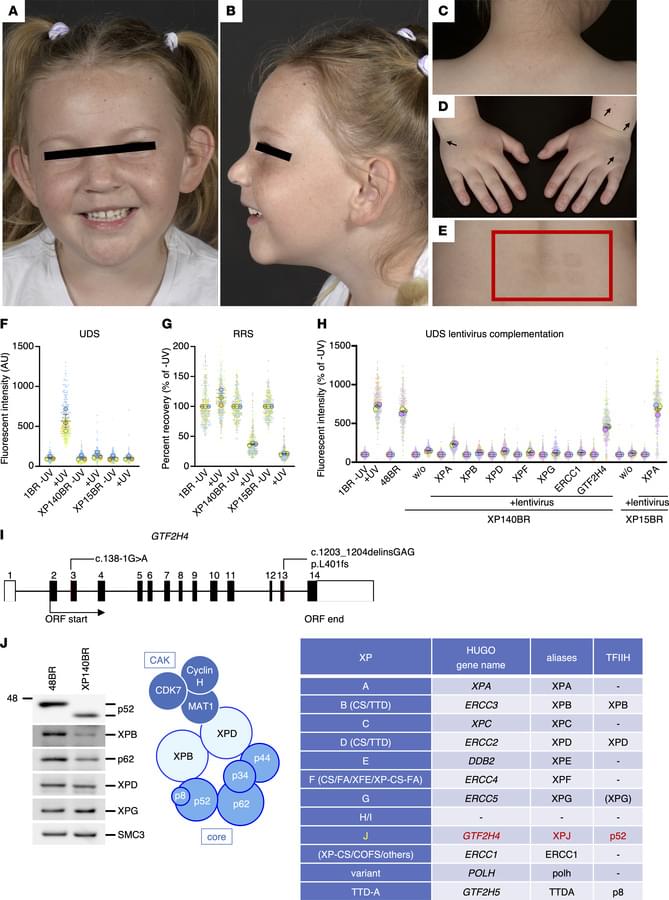
Fassihi et al. and Nakazawa et al. report on the C-terminal deletion of transcription factor TFIIH-p52 subunit as a cause of xeroderma pigmentosum.
The cover art was created using PyMOL; TFIIH-p52 subunit (blue; C-terminus in white). Image credit: Keiko Itano.
https://www.jci.org/articles/view/195731 https://www.jci.org/articles/view/195732
1National Xeroderma Pigmentosum Service, Rare Disease Centre, Guy’s and St Thomas’ NHS Trust, London, United Kingdom.
2Department of Molecular Genetics, Nagoya University Graduate School of Medicine, Nagoya, Japan.

A San Diego biotech has set out to solve the formula for best-in-class antibody-drug conjugates, raising $120 million with support from Big Pharma Merck & Co. to fuel its efforts.
Solve Therapeutics’ new fundraise was led by oncology-focused VC Yosemite, with participation from new investors Abingworth and Merck, plus existing investors Alexandria Venture Investments and Citadel’s Surveyor Capital, among others.