Researchers say a new blood test that analyzes protein biomarkers has shown promise in detecting 18 types of cancer in their early stages.
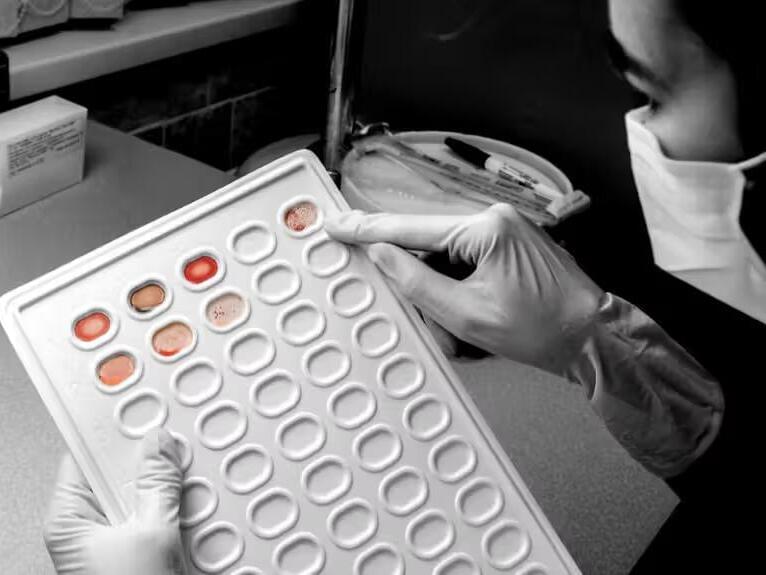


A NEW “pill-on-a-thread” sponge could halve oesophageal cancer deaths in Britain, researchers say.
The new tech quickly tests for Barrett’s oesophagus — a heartburn-causing condition that can lead to cancer.
Patients swallow the capsule containing a sponge, which dissolves in the stomach and expands to the size of a 50p coin before being dragged back up the throat, collecting cells.
Non-personalized content and ads are influenced by things like the content you’re currently viewing and your location (ad serving is based on general location). Personalized content and ads can also include things like video recommendations, a customized YouTube homepage, and tailored ads based on past activity, like the videos you watch and the things you search for on YouTube. We also use cookies and data to tailor the experience to be age-appropriate, if relevant.
Select “More options” to see additional information, including details about managing your privacy settings. You can also visit g.co/privacytools at any time.

Big discovery on the patterns of evolution and how it’ll change medicine and even potentially climate change and synthetic biology.
The experts meticulously analyzed the pangenome — a complete set of genes within a species. By deploying a machine learning technique known as Random Forest, and processing data from 2,500 complete genomes of a single bacterial species, the team embarked on a journey to unravel the mysteries of evolutionary predictability.
“The implications of this research are nothing short of revolutionary,” said Professor McInerney, the lead author of the study.
“By demonstrating that evolution is not as random as we once thought, we’ve opened the door to an array of possibilities in synthetic biology, medicine, and environmental science.”

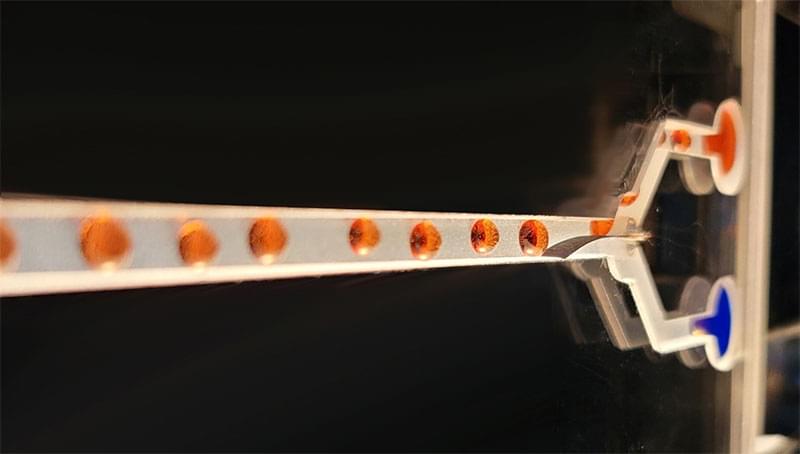
Soft robots, medical devices, and wearable devices have permeated our daily lives. KAIST (Korea Advanced Institute of Science and Technology) researchers have developed a fluid switch using ionic polymer artificial muscles that operates at ultra-low power and produces a force 34 times greater than its weight. Fluid switches control fluid flow, causing the fluid to flow in a specific direction to invoke various movements.
KAIST announced on the 4th of January that a research team under Professor IlKwon Oh from the Department of Mechanical Engineering has developed a soft fluidic switch that operates at ultra-low voltage and can be used in narrow spaces.
The results have been published in Science Advances (“Polysulfonated Covalent Organic Framework as Active Electrode Host for Mobile Cation Guests in Electrochemical Soft Actuator”).

Ichthyosis refers to a group of skin disorders that lead to dry, itchy skin that appears scaly, rough, and red. Your health care provider may be able to diagnose ichthyosis with a genetic test that detects the mutated gene usually from a blood sample or a swab from the mouth. Find out more:
What is ichthyosis? It is a disorder that causes dry, thickened skin that may look similar to fish scales.

In the largest study of its kind, scientists report how combining health data with whole genome sequence (WGS) data in patients with cancer can help doctors provide more tailored care for their patients.
The research, published in Nature Medicine, shows that linking WGS data to real-world clinical data can identify changes in cancer DNA that may be relevant for an individual patient’s care, for example by helping identify what treatment might work best for them based on their cancer.
The study, led by Genomics England, NHS England, Queen Mary University of London, Guy’s and St Thomas’ NHS Foundation Trust and the University of Westminster, analyzed data covering over 30 types of solid tumors collected from more than 13,000 participants with cancer in the 100,000 Genomes Project. By looking at the genomic data alongside routine clinical data collected from participants over a 5-year period, such as hospital visits and the type of treatment they received, scientists were able to find specific genetic changes in the cancer associated with better or worse survival rates and improved patient outcomes.
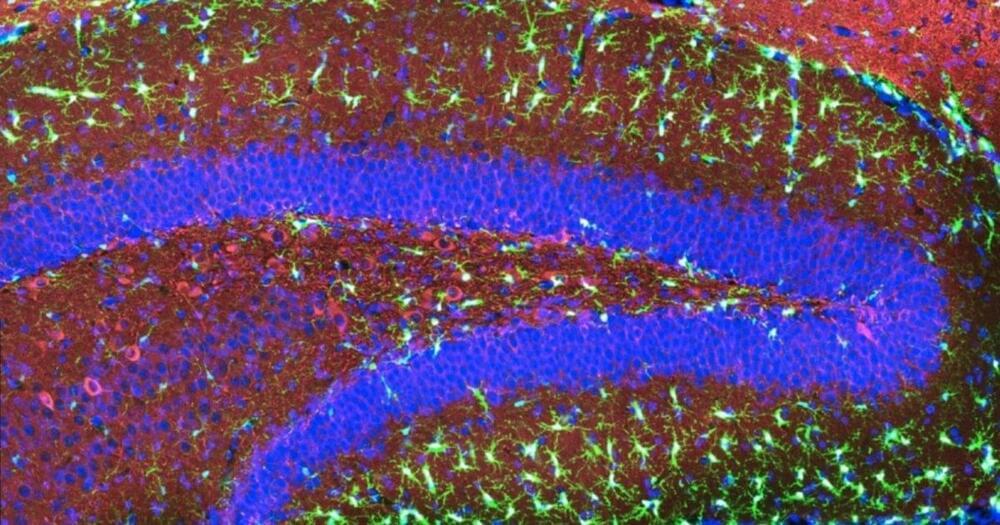

Rapid-acting antidepressants, including ketamine, scopolamine and psilocybin, have been found to have immediate and lasting positive effects on mood in patients with major depressive disorder but how these effects arise is unknown. New research led by the University of Bristol explored their neuropsychological effects and found that all three of these drugs can modulate affective biases associated with learning and memory.
The paper, published in Science Translational Medicine was carried out in collaboration with researchers at Compass Pathways, Boehringer Ingelheim, and the University of Cambridge.
Negative affective biases are a core feature of major depressive disorder. Affective biases occur when emotions alter how the brain processes information and negative affective biases are thought to contribute to the development and continuation of depressed mood.