The surge of treatment-resistant microbes is happening in real time in health care settings across the U.S. Knowing how to deal with it is essential.



“Geometry is destiny”
How a simple cell produces remarkably complex behavior, all without a nervous system.
Combining a deep curiosity and “recreational biology,” Stanford researchers have discovered how a simple cell produces remarkably complex behavior, all without a nervous system. It’s origami, they say.

Strange underwater icicles form in the Earth’s coldest regions and freeze living organisms in place.

Collections of mobile, interacting objects—flocks of birds, colonies of bacterial, or teams of robots—can sometimes behave like solid materials, executing organized rotations or gliding coherently in one direction. But why such systems display one kind of collective organization rather than another has remained unclear. Now researchers have developed a theory that can predict the pattern most likely to emerge under specific conditions [1]. The theory, they hope, may be of use in designing living and artificial materials that can autonomously adapt to their environment.
An “active material” is any system made up of interacting objects able to move under their own power, such as animals, cells, or robots. In so-called active solids, a subset of active materials, strong cohesion between neighboring elements makes the collective act somewhat like a solid. Examples include clusters of certain cell types and networks of robots with rigid connections.
Active solids can display several kinds of collective, organized motion, says Claudio Hernández-López, a PhD student at the École Normale Supérieure and Sorbonne University in France. For example, researchers have observed both coherent rotations and coherent translations in collections of microbes from the phylum Placozoa. Existing theories, however, fail to explain pattern selection—why, if several patterns are possible, does one pattern of behavior emerge rather than another?

“… living systems evolve to exploit any aspect of physics that enables exploration of all possible ‘fitness landscapes’.”
Indeed!
In 1990, within the intellectual haven of Haverford College, I embarked on a transformative academic journey into biophysics – the captivating intersection of physics and biology.
It was during this time that I delved into the tantalising notion of quantum mechanics operating within living organisms.
Unbeknownst to me, this exploration would etch an enduring imprint on my scientific voyage, kindling a lifelong fascination with biophysics. Ultimately, I charted my research course in quantum cosmology, but the echoes of biophysics persisted.

As humanity travels back to the Moon in the next few years and potentially Mars in the next decade, how much of a role should planetary protection play regarding the safeguarding of these worlds? This is what a recent study published in Space Policy hopes to address as a team of international researchers discuss prioritizing planetary protection and sustainability could not only aid in space exploration but also sustainability on Earth, as well.
For the study, the researchers propose the expansion of current planetary protection policies to help safeguard against security, orbital debris, and crowding, as current policies only focus on preventing biological contamination from human activities. While biological contamination might not be a concern on the Moon given it lacks the necessary conditions to support life, the planet Mars is hypothesized to have once possessed microbial life deep in its ancient past and could potentially be hosting life beneath its surface.
“Sustainability must become a core principle of human space exploration,” said Dr. Dimitra Atri, who is an investigator in the Center for Astrophysics and Space Science at NYU Abu Dhabi and lead author of the study. “Just as we view climate change as the great challenge facing our terrestrial human society, the space community should begin to address space sustainability with the same urgency.”
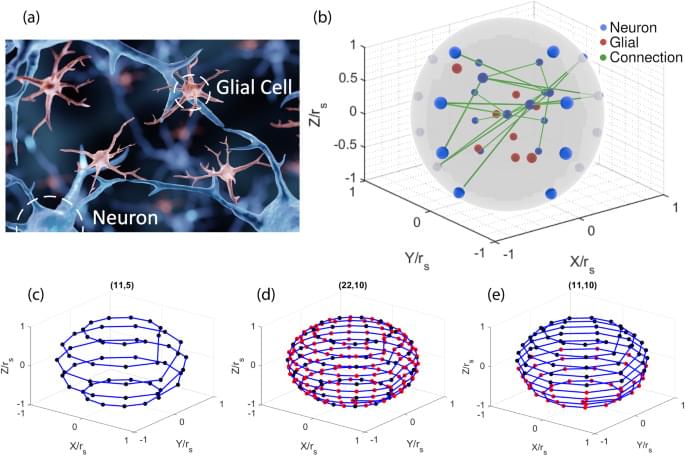
In artificial neural networks, many models are trained for a narrow task using a specific dataset. They face difficulties in solving problems that include dynamic input/output data types and changing objective functions. Whenever the input/output tensor dimension or the data type is modified, the machine learning models need to be rebuilt and subsequently retrained from scratch. Furthermore, many machine learning algorithms that are trained for a specific objective, such as classification, may perform poorly at other tasks, such as reinforcement learning or quantification.
Even if the input/output dimensions and the objective functions remain constant, the algorithms do not generalize well across different datasets. For example, a neural network trained on classifying cats and dogs does not perform well on classifying humans and horses despite both of the datasets having the exact same image input1. Moreover, neural networks are highly susceptible to adversarial attacks2. A small deviation from the training dataset, such as changing one pixel, could cause the neural network to have significantly worse performance. This problem is known as the generalization problem3, and the field of transfer learning can help to solve it.
Transfer learning4,5,6,7,8,9,10 solves the problems presented above by allowing knowledge transfer from one neural network to another. A common way to use supervised transfer learning is obtaining a large pre-trained neural network and retraining it for a different but closely related problem. This significantly reduces training time and allows the model to be trained on a less powerful computer. Many researchers used pre-trained neural networks such as ResNet-5011 and retrained them to classify malicious software12,13,14,15. Another application of transfer learning is tackling the generalization problem, where the testing dataset is completely different from the training dataset. For example, every human has unique electroencephalography (EEG) signals due to them having distinctive brain structures. Transfer learning solves the generalization problem by pretraining on a general population EEG dataset and retraining the model for a specific patient16,17,18,19,20. As a result, the neural network is dynamically tailored for a specific person and can interpret their specific EEG signals properly. Labeling large datasets by hand is tedious and time-consuming. In semi-supervised transfer learning21,22,23,24, either the source dataset or the target dataset is unlabeled. That way, the neural networks can self-learn which pieces of information to extract and process without many labels.
Chapters: 0:00 Colin Wright Highlights 0:48 Colin Wright: A Horrible Person, A Transphobe? 3:43 Did This Piss Colin Off? 6:03 Humans Will Always Do Magical Thinking 8:32 If We Stand Up Together… 9:48 The Fundamental Misunderstanding / Fish 12:48 What Activists Get Wrong (Secondary Characteristics) 15:48 The ‘True’ Hermaphrodite 17:48 Is There A Male or Female Brain? 21:48 Judith Butler’s Contradiction 24:48 Individual Liberty 27:48 Young Girls & Older Men 30:48 Cross-Dressers Getting Aroused 34:18 How Sex Is Determined In Nature 37:38 Why Do Men Have Nipples? 38:58 Why Don’t Testicles Have Rib Cages? 40:18 Creationism vs Evolution (Joe Rogan) 44:18 Alex Jones & Gay Frogs 45:08 What Does ‘Theory’ of Evolution Mean? 48:08 Other Competing Theories? 51:28 Faith vs Science 53:48 Danger of Reality Denial 57:43 A Heretic Colin Admires.