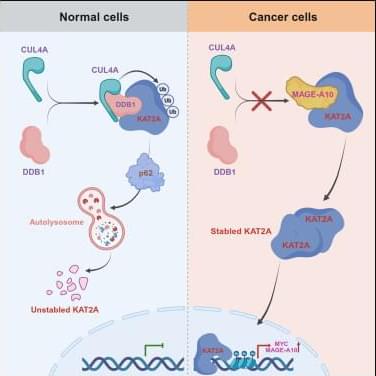
Fu et al. show that MAGE-A10 drives tumorigenesis by blocking autophagic degradation of KAT2A/KAT2B and enhancing histone acetylation.

Join us on Patreon! https://www.patreon.com/MichaelLustgartenPhD
Discount Links/Affiliates:
Blood testing (where I get the majority of my labs): https://www.ultalabtests.com/partners/michaellustgarten.
At-Home Metabolomics: https://www.iollo.com?ref=michael-lustgarten.
Use Code: CONQUERAGING At Checkout.
Clearly Filtered Water Filter: https://get.aspr.app/SHoPY
Epigenetic, Telomere Testing: https://trudiagnostic.com/?irclickid=U-s3Ii2r7xyIU-LSYLyQdQ6…M0&irgwc=1
Use Code: CONQUERAGING
NAD+ Quantification: https://www.jinfiniti.com/intracellular-nad-test/

Daley and Mout added that the team is excited that the approach can guide T cells to tumors, stimulate their cancer cell-killing abilities, and overcome immune suppression by the tumor microenvironment.
Mout, who trained in the lab of Nobel Prize-winning co-author and Rosetta creator David Baker, is especially enthusiastic about the technology’s far-reaching potential.
“Our goal is to develop next-generation immunotherapies and cancer vaccines,” he said.
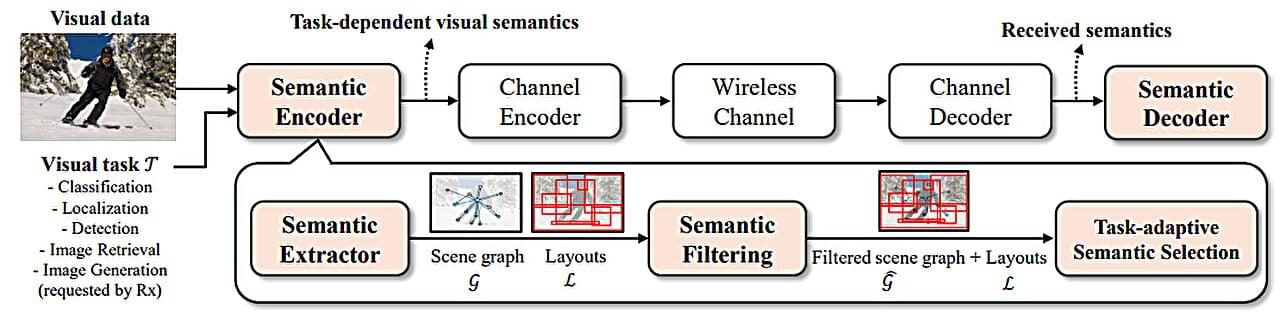
A new AI-driven technology developed by researchers at UNIST promises to significantly reduce data transmission loads during image transfer, paving the way for advancements in autonomous vehicles, remote surgery and diagnostics, and real-time metaverse rendering—applications that demand rapid, large-scale visual data exchange without delay.
Led by Professor Sung Whan Yoon from the Graduate School of Artificial Intelligence at UNIST, the research team developed Task-Adaptive Semantic Communication, an innovative wireless image transmission method that selectively transmits only the most essential semantic information relevant to the specific task. Their study is published in the IEEE Journal on Selected Areas in Communications.
Current wireless image transmission methods compress entire images without considering their underlying semantic structures—such as objects, layout, and relationships—resulting in bandwidth limitations and transmission delays that hinder real-time high-resolution image sharing.
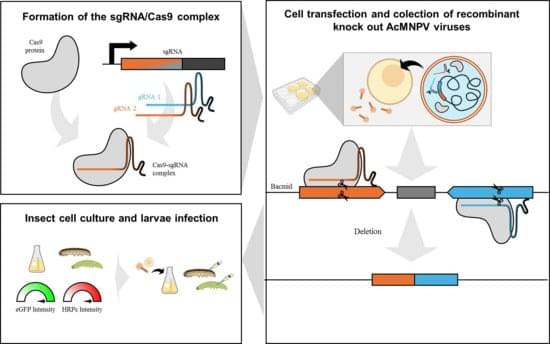
The CRISPR/Cas9 system is a powerful genome-editing tool that is applied in baculovirus engineering. In this study, we present the first report of the AcMNPV genome deletions for bioproduction purposes, using a dual single-guide RNA (sgRNA) CRISPR/Cas9 approach. We used this method to remove nonessential genes for the budded virus and boost recombinant protein yields when applied as BEVS. We show that the co-delivery of two distinct ribonucleoprotein (RNP) complexes, each assembled with a sgRNA and Cas9, into Sf9 insect cells efficiently generated deletions of fragments containing tandem genes in the genome. To evaluate the potential of this method, we assessed the expression of two model proteins, eGFP and HRPc, in insect cells and larvae. The gene deletions had diverse effects on protein expression: some significantly enhanced it while others reduced production.