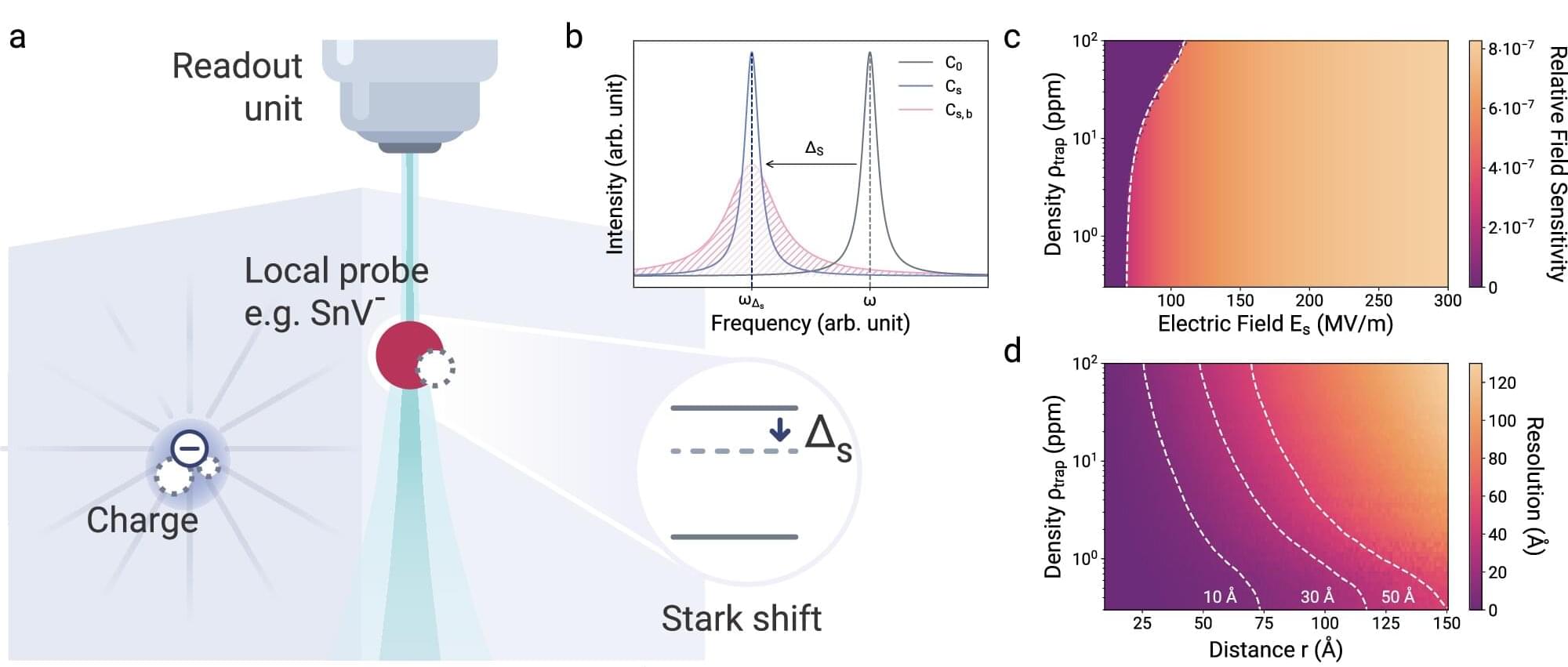
From computer chips to quantum dots—technological platforms were only made possible thanks to a detailed understanding of the used solid-state materials, such as silicon or more complex semiconductor materials. This understanding also includes being able to identify and control irregularities in the crystal lattice of such materials.
If, for example, an atom is missing in the lattice structure of the crystals, a single electron and thus an electric charge can become trapped there. Such charge traps generate electromagnetic noise that limits the functionality of these materials. However, it is extremely difficult to locate these charge traps on an atomic scale.
Researchers from the “Integrated Quantum Photonics” group at the Department of Physics at Humboldt-Universität zu Berlin (HU) and the “Joint Lab Diamond Nanophotonics” at the Ferdinand-Braun-Institut, led by Prof. Dr. Tim Schröder, have developed a new sensor that can detect such individual electrical charges more precisely than ever before.