Traditional drug development methods involve identifying a target protein (e.g., a cancer cell receptor) that causes disease, and then searching through countless molecular candidates (potential drugs) that could bind to that protein and block its function. This process is costly, time-consuming, and has a low success rate.
KAIST researchers have developed an AI model that, using only information about the target protein, can design optimal drug candidates without any prior molecular data—opening up new possibilities for drug discovery. The research is published in the journal Advanced Science.
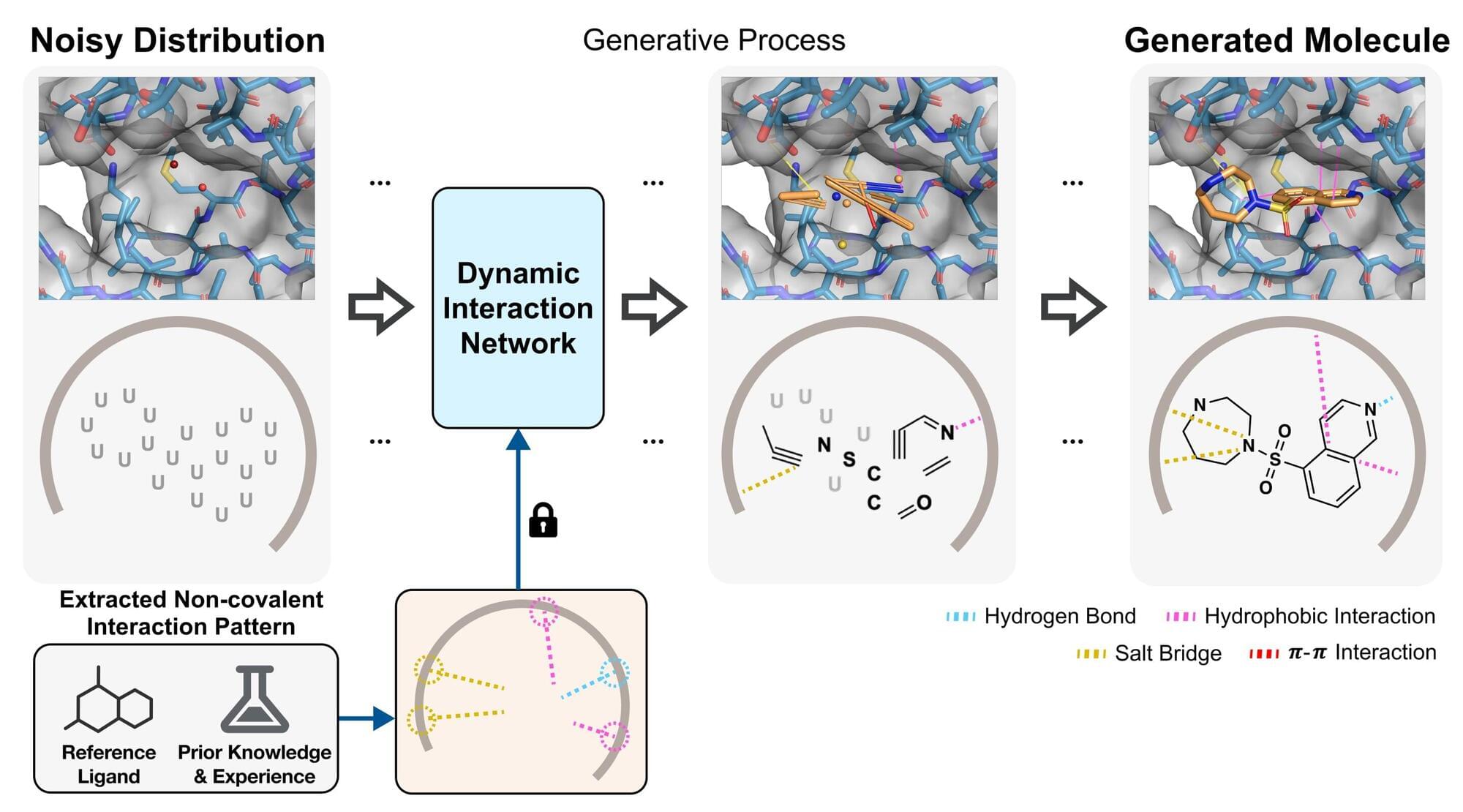
The research team led by Professor Woo Youn Kim in the Department of Chemistry has developed an AI model named BInD (Bond and Interaction-generating Diffusion model), which can design and optimize drug candidate molecules tailored to a protein’s structure alone—without needing prior information about binding molecules. The model also predicts the binding mechanism (non-covalent interactions) between the drug and the target protein.