NRG Energy announced Monday that they plan to acquire 18 natural gas facilities across nine states, including Texas.
Personal Perspective: As AI evolves toward possible consciousness, how we engage with it today could shape the nature of a future shared with a new intelligent species.
Could light’s behavior in the double-slit experiment be explained without waves? Discover the groundbreaking “dark photon” theory that’s turning quantum physics on its head. Dive into how bright and dark photon states could rewrite our understanding of interference, measurement, and reality itself. Watch now!
Paper link: https://journals.aps.org/prl/abstract… 00:00 Introduction 01:17 Rethinking the Double-Slit — Not a Wave After All? 04:10 Bright vs. Dark — Redefining Reality Through Detection 07:10 Implications and Related Discoveries — From Theory to Possibility 10:04 Outro 10:26 Enjoy MUSIC TITLE : Starlight Harmonies MUSIC LINK : https://pixabay.com/music/pulses-star… Visit our website for up-to-the-minute updates: www.nasaspacenews.com Follow us Facebook: / nasaspacenews Twitter:
/ spacenewsnasa Join this channel to get access to these perks:
/ @nasaspacenewsagency #NSN #NASA #Astronomy#DarkPhoton #QuantumPhysics #DoubleSlitExperiment #QuantumMechanics #PhysicsBreakthrough #QuantumTheory #BrightPhotons #DarkPhotons #QuantumInterference #QuantumMeasurement #MaxPlanckInstitute #GerhardRempe #QuantumSuperposition #WaveParticleDuality #QuantumLight #QuantumEntanglement #QuantumReality #QuantumRevolution #ScienceNews #LightInterference #Photons #NewPhysics #PhysicsTheory #QuantumMystery #QuantumWorld #QuantumExplanation #QuantumScience #QuantumExperiments #QuantumTech #InvisibleParticles.
Chapters:
00:00 Introduction.
01:17 Rethinking the Double-Slit — Not a Wave After All?
04:10 Bright vs. Dark — Redefining Reality Through Detection.
07:10 Implications and Related Discoveries — From Theory to Possibility.
10:04 Outro.
10:26 Enjoy.
MUSIC TITLE : Starlight Harmonies.
MUSIC LINK : https://pixabay.com/music/pulses-star…
Visit our website for up-to-the-minute updates:
www.nasaspacenews.com.
Follow us.
#neuroscience #biology #artificialintelligence #phenomenology #science In Philosophy Babble — The Great Minds SeriesIn this instal…
Cisco has unveiled a prototype quantum entanglement chip and opened a dedicated quantum lab, advancing its quantum internet strategy.
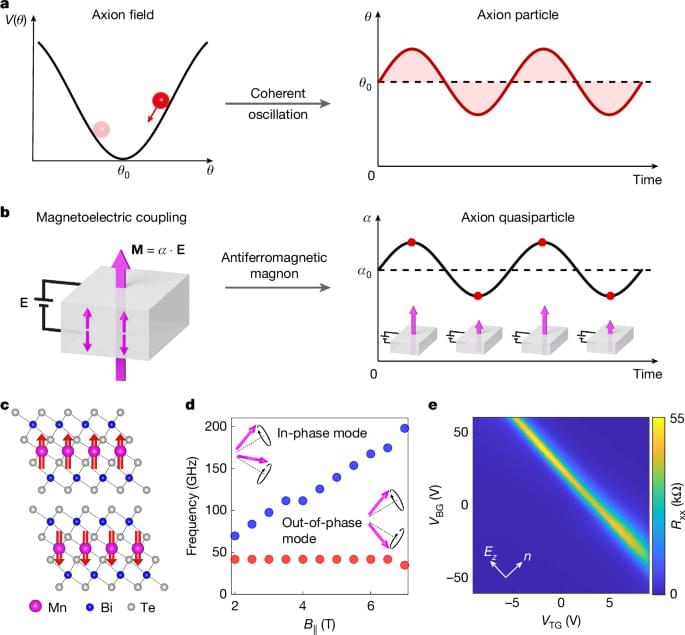
The dynamical axion quasiparticle, which is directly analogous to the hypothetical fundamental axion particle, is observed in two-dimensional MnBi2Te4, and has implications for quantum chromodynamics, cosmology and string theory.
Walgreens is expanding the number of its retail stores served by its micro-fulfillment centers as it works to turn itself around and prepares to go private.
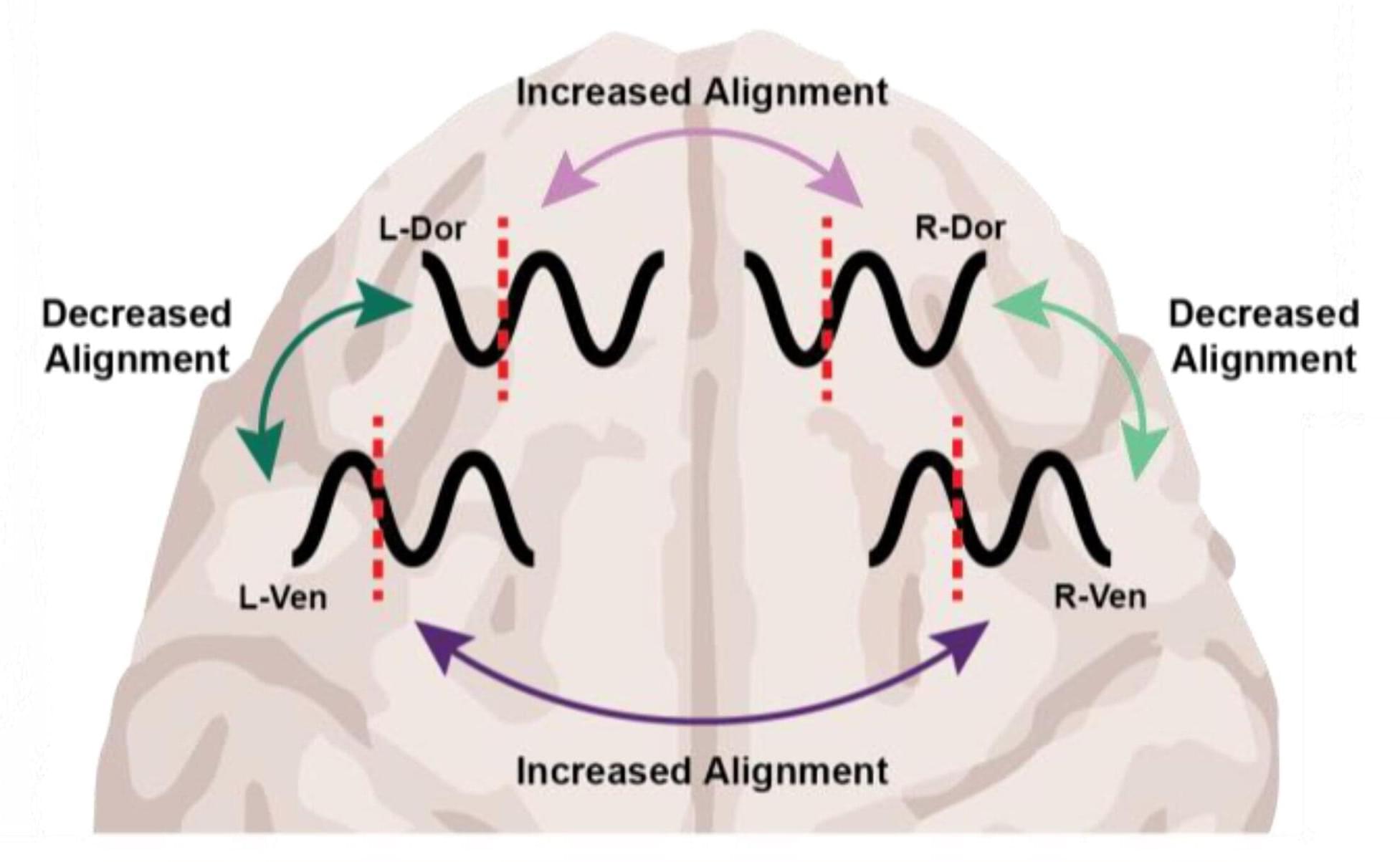
At the level of molecules and cells, ketamine and dexmedetomidine work very differently, but in the operating room they do the same exact thing: anesthetize the patient. By demonstrating how these distinct drugs achieve the same result, a new study in animals by neuroscientists at The Picower Institute for Learning and Memory at MIT identifies a potential signature of unconsciousness that is readily measurable to improve anesthesiology care.
What the two drugs have in common, the researchers discovered, is the way they push around brain waves, which are produced by the collective electrical activity of neurons.
When brain waves are in phase, meaning the peaks and valleys of the waves are aligned, local groups of neurons in the brain’s cortex can share information to produce conscious cognitive functions such as attention, perception and reasoning, said Picower Professor Earl K. Miller, senior author of the new study in Cell Reports. When brain waves fall out of phase, local communications, and therefore functions, fall apart, producing unconsciousness.
MIT physicists have captured the first images of individual atoms freely interacting in space. The pictures reveal correlations among the “free-range” particles that until now were predicted but never directly observed. Their findings, appearing today in the journal Physical Review Letters, will help scientists visualize never-before-seen quantum phenomena in real space.
The images were taken using a technique developed by the team that first allows a cloud of atoms to move and interact freely. The researchers then turn on a lattice of light that briefly freezes the atoms in their tracks, and apply finely tuned lasers to quickly illuminate the suspended atoms, creating a picture of their positions before the atoms naturally dissipate.
The physicists applied the technique to visualize clouds of different types of atoms, and snapped a number of imaging firsts. The researchers directly observed atoms known as “bosons,” which bunched up in a quantum phenomenon to form a wave. They also captured atoms known as “fermions” in the act of pairing up in free space — a key mechanism that enables superconductivity.
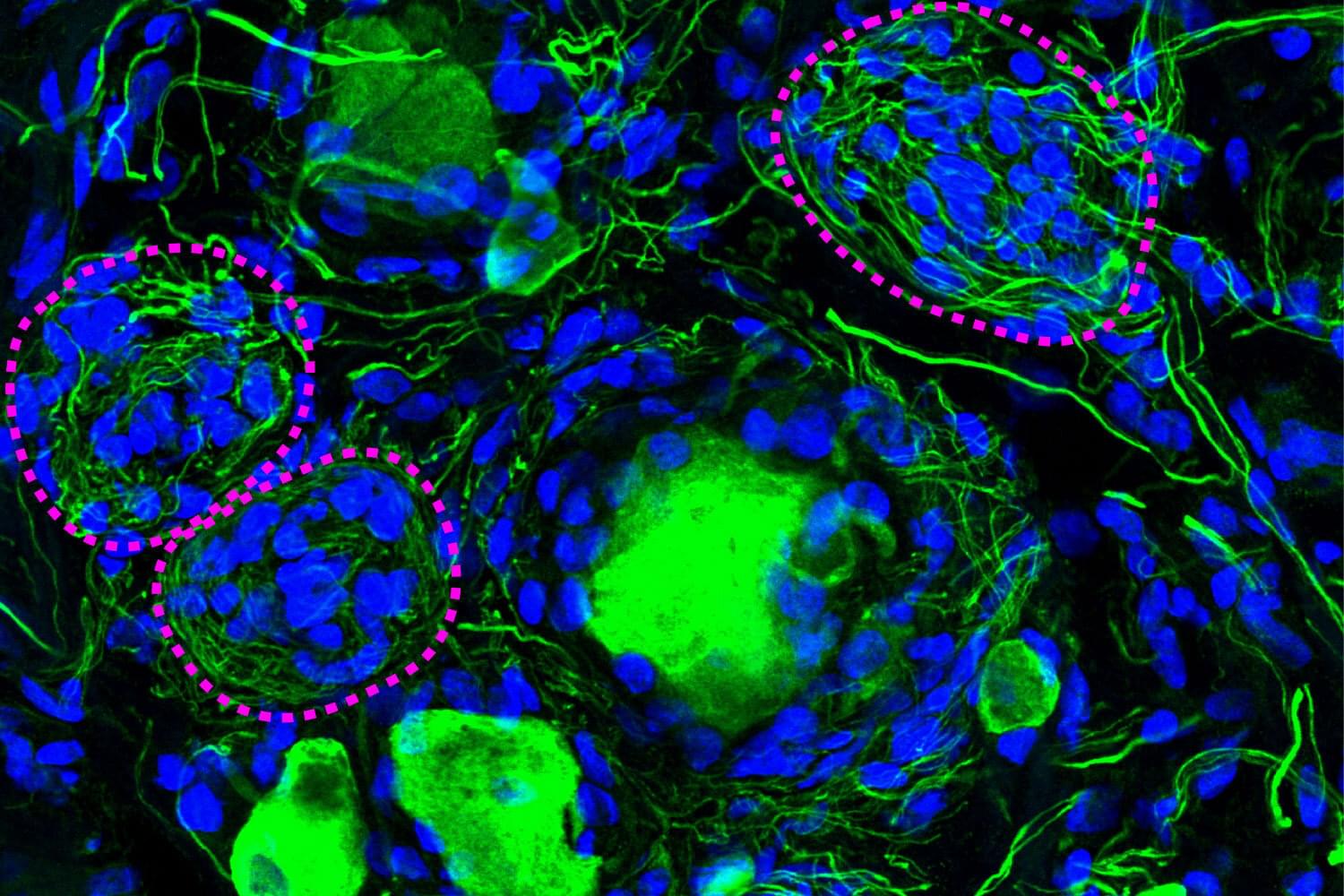
A phenomenon largely ignored since its discovery 100 years ago appears to be a crucial component of diabetic pain, according to new research from The University of Texas at Dallas’s Center for Advanced Pain Studies (CAPS).
Findings from a new study published in Nature Communications suggest that cell clusters called Nageotte nodules are a strong indicator of nerve cell death in human sensory ganglia. These could prove to be a target for drugs that would protect these nerves or help manage diabetic neuropathy.
“The key finding of our study is really a new view of diabetic neuropathic pain,” said Dr. Ted Price, Ashbel Smith Professor of neuroscience in the School of Behavioral and Brain Sciences, CAPS director and co-corresponding author of the study. “We believe our data demonstrate that neurodegeneration in the dorsal root ganglion is a critical facet of the disease—which should really force us to think about the disease in a new and urgent way.”