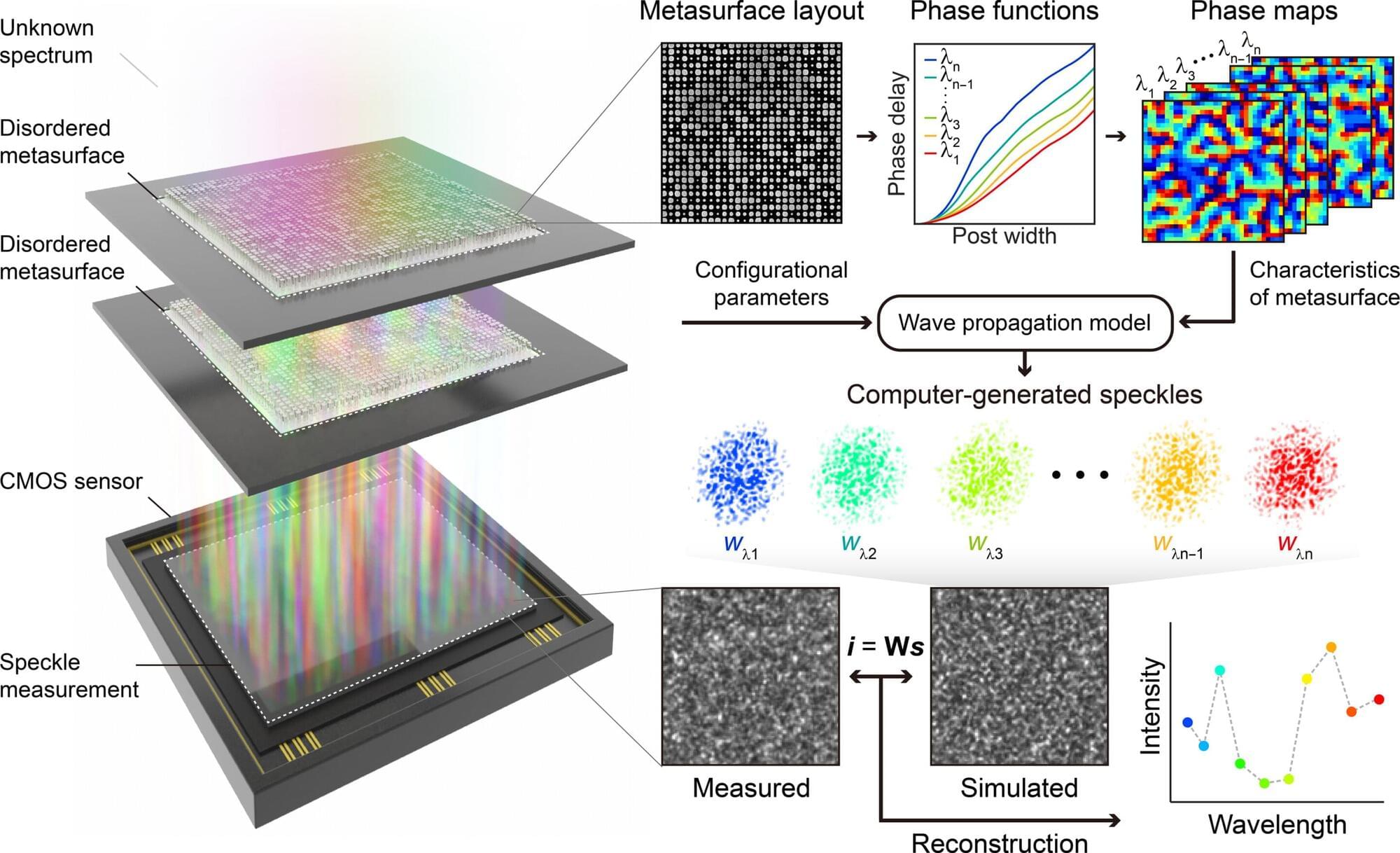
Color, as the way light’s wavelength is perceived by the human eye, goes beyond a simple aesthetic element, containing important scientific information like a substance’s composition or state.
Spectrometers are optical devices that analyze material properties by decomposing light into its constituent wavelengths, and they are widely used in various scientific and industrial fields, including material analysis, chemical component detection, and life science research.
Existing high-resolution spectrometers were large and complex, making them difficult for widespread daily use. However, thanks to the ultra-compact, high-resolution spectrometer developed by KAIST researchers, it is now expected that light’s color information can be utilized even within smartphones or wearable devices.