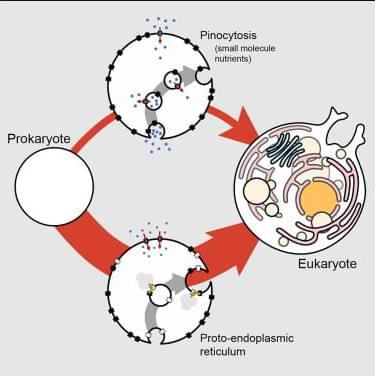
Schavemaker and Lynch derived functions that relate the cell biology of endomembranes to cellular fitness. Applied to the pinocytosis of small-molecule nutrients and the insertion of membrane proteins by a proto-endoplasmic reticulum, the proto-endoplasmic reticulum is revealed to be the more likely path to complex endomembranes in the origin of eukaryotes.
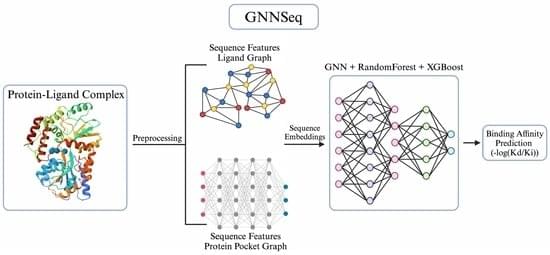
Background/Objectives: Accurately predicting protein–ligand binding affinity is essential in drug discovery for identifying effective compounds. While existing sequence-based machine learning models for binding affinity prediction have shown potential, they lack accuracy and robustness in pattern recognition, which limits their generalizability across diverse and novel binding complexes. To overcome these limitations, we developed GNNSeq, a novel hybrid machine learning model that integrates a Graph Neural Network (GNN) with Random Forest (RF) and XGBoost. Methods: GNNSeq predicts ligand binding affinity by extracting molecular characteristics and sequence patterns from protein and ligand sequences. The fully optimized GNNSeq model was trained and tested on subsets of the PDBbind dataset. The novelty of GNNSeq lies in its exclusive reliance on sequence features, a hybrid GNN framework, and an optimized kernel-based context-switching design. By relying exclusively on sequence features, GNNSeq eliminates the need for pre-docked complexes or high-quality structural data, allowing for accurate binding affinity predictions even when interaction-based or structural information is unavailable. The integration of GNN, XGBoost, and RF improves GNNSeq performance by hierarchical sequence learning, handling complex feature interactions, reducing variance, and forming a robust ensemble that improves predictions and mitigates overfitting. The GNNSeq unique kernel-based context switching scheme optimizes model efficiency and runtime, dynamically adjusts feature weighting between sequence and basic structural information, and improves predictive accuracy and model generalization. Results: In benchmarking, GNNSeq performed comparably to several existing sequence-based models and achieved a Pearson correlation coefficient (PCC) of 0.784 on the PDBbind v.2020 refined set and 0.84 on the PDBbind v.2016 core set. During external validation with the DUDE-Z v.2023.06.20 dataset, GNNSeq attained an average area under the curve (AUC) of 0.74, demonstrating its ability to distinguish active ligands from decoys across diverse ligand–receptor pairs. To further evaluate its performance, we combined GNNSeq with two additional specialized models that integrate structural and protein–ligand interaction features. When tested on a curated set of well-characterized drug–target complexes, the hybrid models achieved an average PCC of 0.89, with the top-performing model reaching a PCC of 0.97. GNNSeq was designed with a strong emphasis on computational efficiency, training on 5000+ complexes in 1 h and 32 min, with real-time affinity predictions for test complexes. Conclusions: GNNSeq provides an efficient and scalable approach for binding affinity prediction, offering improved accuracy and generalizability while enabling large-scale virtual screening and cost-effective hit identification. GNNSeq is publicly available in a server-based graphical user interface (GUI) format.
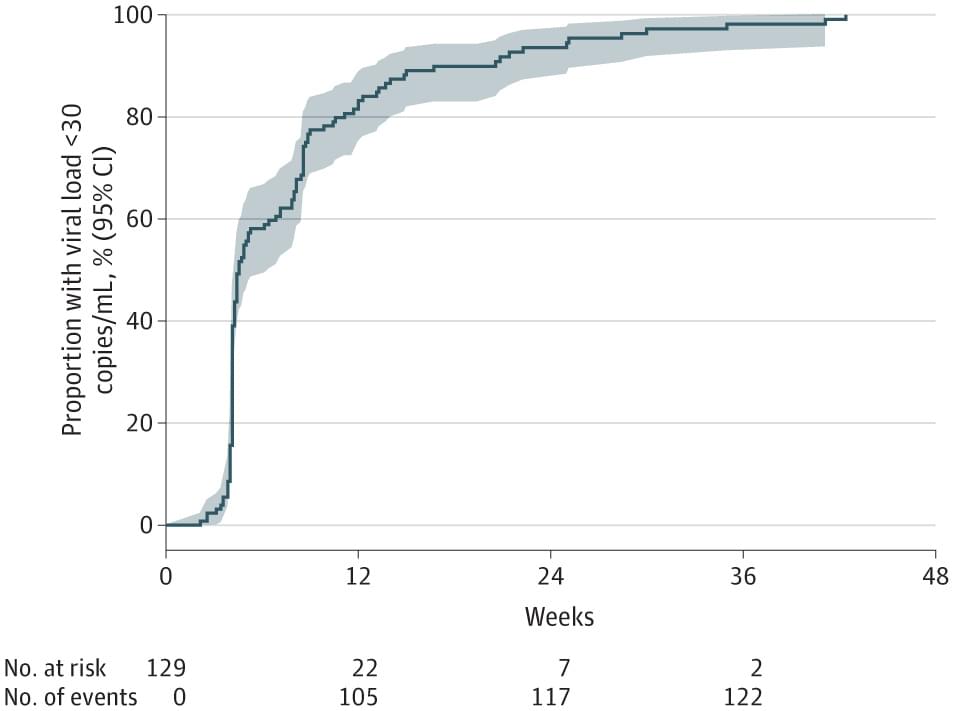
This study uses electronic medical record data to compare 48-week viral load outcomes after starting long-acting antiretroviral therapy among people in the US with HIV with or without viremia from January 2021 through September 2024.
Media conglomerate Ziff Davis claims ChatGPT creator has used their works for AI development.
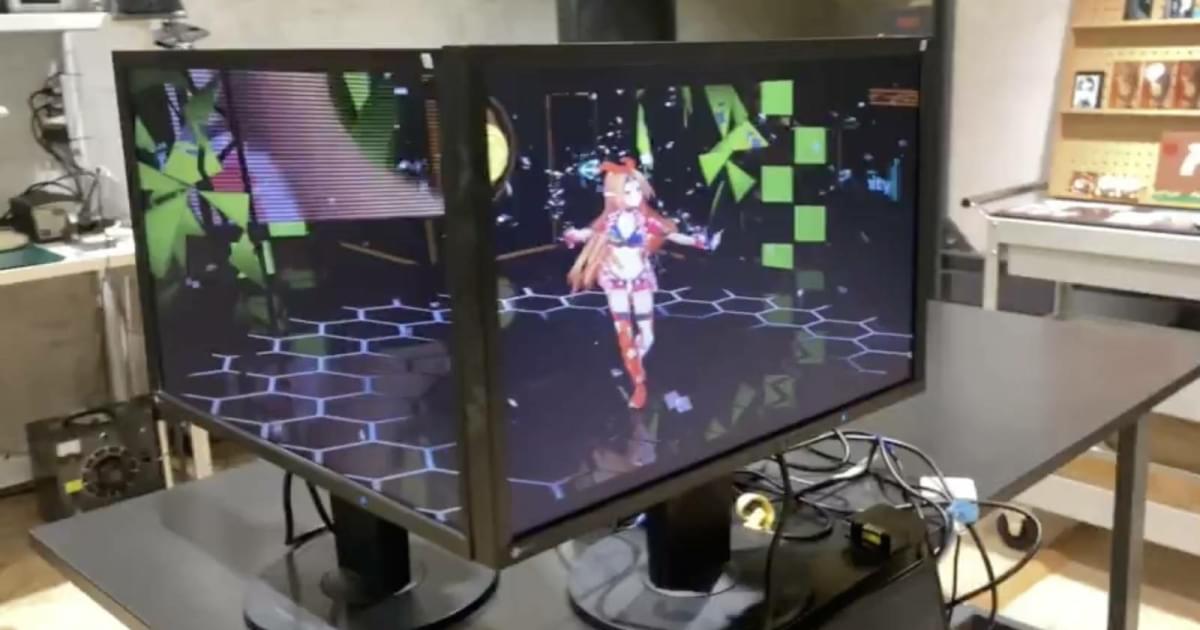
It’s easy to solve a 3×3 Rubik’s cube, says Shantanu Chakrabartty, the Clifford W. Murphy Professor and vice dean for research and graduate education in the McKelvey School of Engineering at Washington University in St. Louis. Just learn and memorize the steps then execute them to arrive at the solution.
Computers are already good at this kind of procedural problem solving. Now, Chakrabartty and his collaborators have developed a tool that can go beyond procedure to discover new solutions to complex optimization problems in logistics to drug discovery.
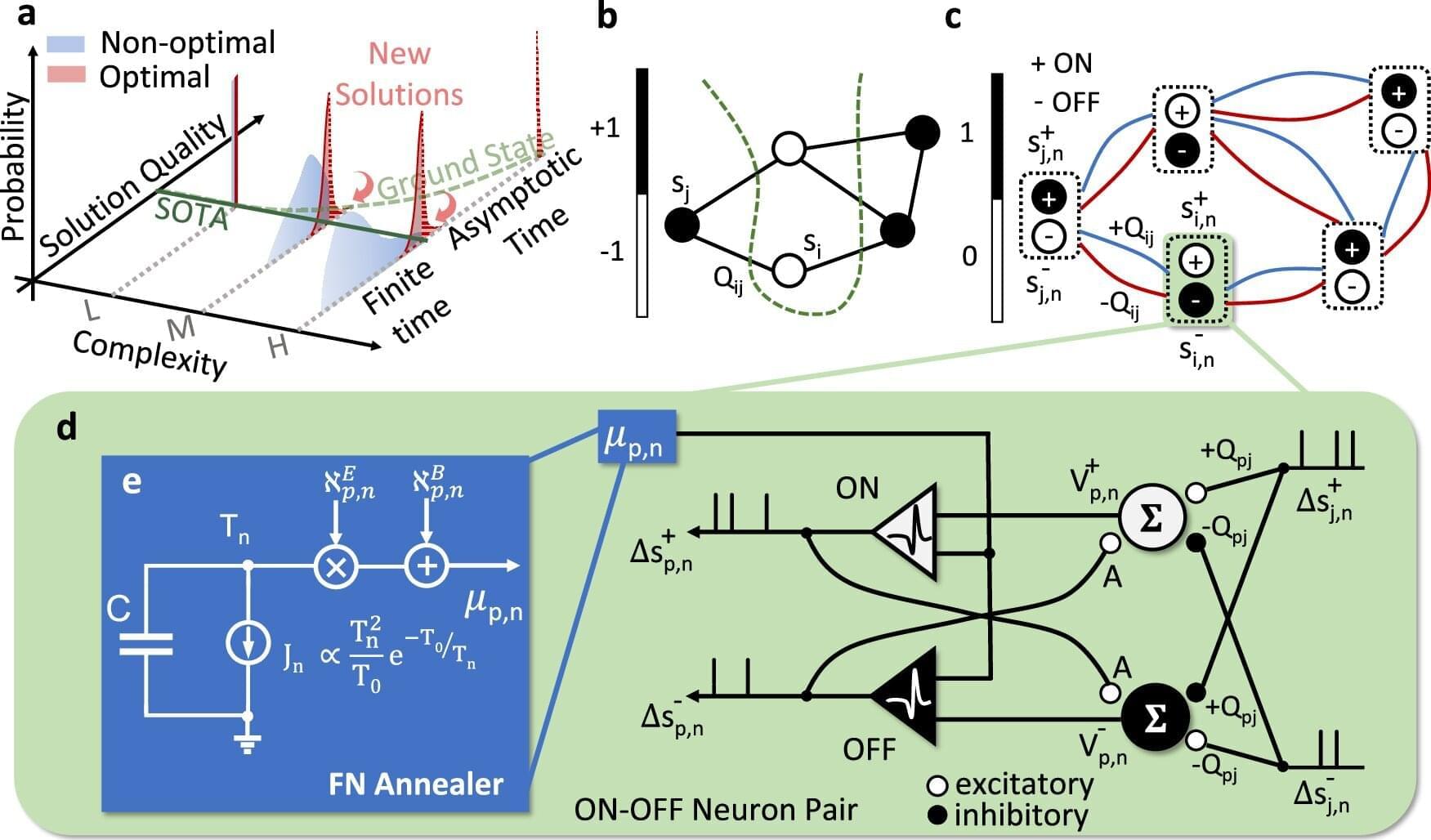
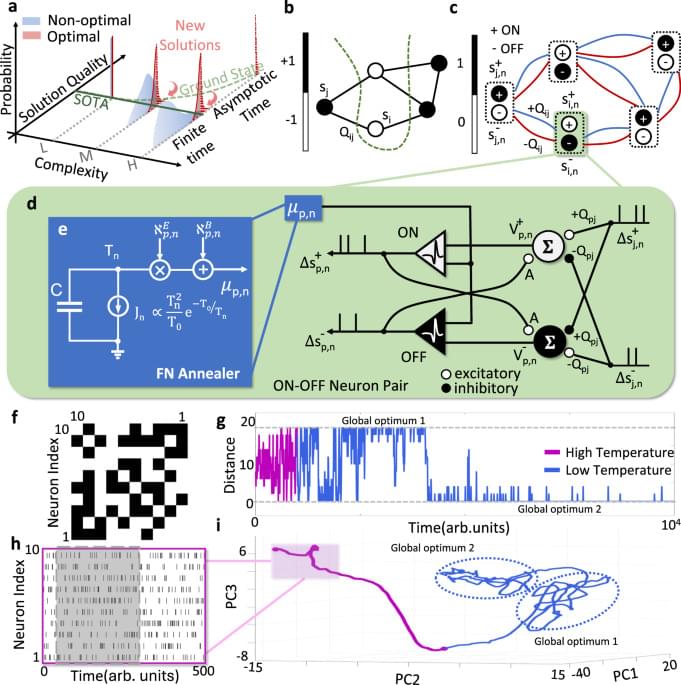
Chakrabartty and his collaborators introduced NeuroSA, a problem-solving neuromorphic architecture modeled on how human neurobiology functions, but that leverages quantum mechanical behavior to find optimal solutions—guaranteed—and find those solutions more reliably than state-of-the-art methods.
Combinatorial Optimization problems can be solved by investigating the ground states of particular Ising models. Here, the authors developed a neuromorphic architecture to ensure asymptotic convergence to the ground state of an Ising problem and to consistently produce high-quality solutions.
A new study highlights a potential role for immune system dysfunction in the onset and progression of Alzheimer’s disease.
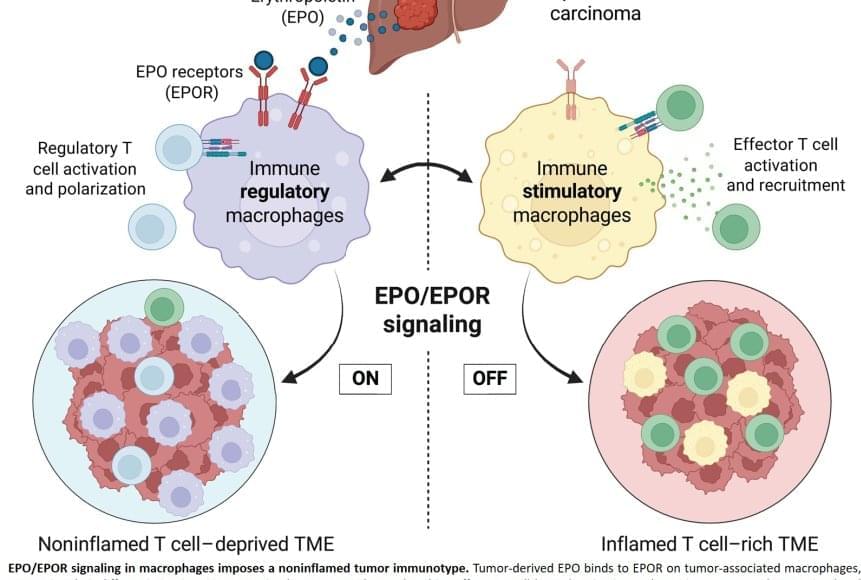
A protein identified nearly 40 years ago for its ability to stimulate the production of red blood cells plays a surprising, critical role in dampening the immune system’s response to cancer.
Blocking the activity of the protein turns formerly “cold,” or immune-resistant, liver tumors in mice into “hot” tumors teeming with cancer-fighting immune cells. When combined with an immunotherapy that further activates these immune cells against the cancer, the treatment led to complete regression of existing liver tumors in most mice. Treated animals lived for the duration of the experiment. In contrast, control animals survived only a few weeks.
“This is a fundamental breakthrough in our understanding of how the immune system is turned off and on in cancer,” said the senior author published the work in Science. “I could not be more excited about this discovery, and I hope treatments that target the mechanism we uncovered will quickly move forward to human trials.”